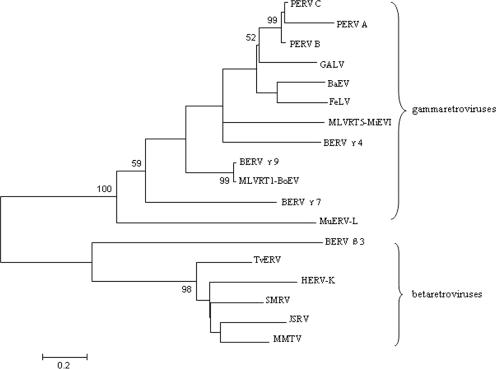
FIG. 2.
Phylogenetic analysis of previously identified retroviruses and BERVs based on the amino acid sequences of the pro/pol region using the neighbor joining method. Numbers at the branch nodes denote the bootstrap values (>50) from 500 replicates. The BERV β and γ families clustered with betaretroviruses (jaagsiekte sheep retrovirus [JSRV], mouse mammary tumor virus [MMTV], simian sarcoma virus [SMRV], Trichosurus vulpecula retrovirus [TvERV], and HERV-K) and gammaretroviruses (PERV-A, PERV-B, PERV-C, gibbon ape leukemia virus [GALV], feline leukemia virus [FeLV], baboon endogenous virus [BaEV], MLVRT1-BoEV, and MLVRT5-MiEVI), respectively. The viruses and GenBank accession numbers of the sequences used in the phylogenetic tree are as follows: JSRV, AAA89182; MMTV, AAA46542; SMRV, AAA66453; TvERV, AAF36395; HERV-K, CAB56603; and murine ERV-L (MuERV-L), CAA73251.