Abstract
The human papillomavirus (HPV) type 16 (HPV16) E6 protein stimulates transcription of the catalytic subunit of telomerase, hTERT, in epithelial cells. It has been reported that binding to the ubiquitin ligase E6AP is required for this E6 activity, with E6 directing E6AP to the hTERT promoter. We previously reported two E6AP binding-defective HPV16 E6 mutations that induced immortalization of human mammary epithelial cells. Because activation of hTERT is proposed to be necessary for epithelial cell immortalization, we sought to further characterize the relationship between E6/E6AP association and telomerase induction. We demonstrate that while these E6 mutants do not bind E6AP, they retain the capability to stimulate the expression of hTERT. Chromatin immunoprecipitation assays confirmed the presence of Myc, wild-type E6, and the E6AP binding-defective E6 mutants, but not E6AP itself, at the endogenous hTERT promoter. Interestingly, an immortalization-defective E6 mutant localized to the hTERT promoter but failed to increase transcription. We conclude that binding to E6AP is not necessary for E6 localization to or activation of the hTERT promoter and that another activity of E6 is involved in hTERT activation.
The best-characterized property of the E6 protein of the cancer-associated human papillomavirus (HPV) is the degradation of p53 mediated by the ubiquitin ligase E6AP (20, 38). Additionally, E6 has several p53-independent activities. HPV type 16 (HPV16) E6 binds to and inhibits the transactivation functions of histone acetyltransferase (HAT) proteins CBP and p300 (36, 51). E6 also interacts with the calcium binding protein E6BP/ERC-55, the Rap GTPase-activating protein E6TP1, and the p53 coactivator hAda3 (8, 15, 27). An important activity of E6 thought to be necessary for epithelial cell immortalization is its ability to increase telomerase activity (16, 26, 35).
Telomerase is a ribonucleoprotein complex that maintains telomere length. Its RNA subunit serves as a template for the synthesis of telomeric DNA, and its catalytic subunit has been shown to be necessary for telomerase enzymatic activity (49). Most somatic cells have no or very low telomerase activity (4). Senescence can be bypassed, and several primary human cell types can be immortalized, by forced expression of the telomerase catalytic subunit hTERT (4, 24, 44). Cellular oncogenes, as well as HPV16 E6, are known to induce hTERT transcription (10, 26, 28, 47). The hTERT core promoter region contains E and GC boxes with recognition sites for transcription factors including Myc, SP-1, and USF (9, 17, 18, 42). Myc activates hTERT, while other factors, such as Mad, Sip1, and Menin, repress hTERT expression (19, 28, 34). Myc and Mad are highly unstable proteins and function by forming dimers with the stable protein Max (3, 12). It has been suggested that the hTERT promoter is differentially regulated by switching binding between Myc/Max and Mad/Max dimers (48). This is supported by the finding that Myc/Max complexes were dominant in immortal cells with high telomerase activity while Mad/Max dimers were abundant in mortal cells (34).
In contrast to Myc, which induces telomerase in both epithelial cells and fibroblasts, HPV16 E6 activation of telomerase is limited to epithelial cells (26). How E6 activates telomerase has not been resolved. E6 mutants unable to induce p53 degradation still increased hTERT expression, implying that telomerase activation by E6 is a p53-independent effect (26, 31). Subsequent reports have proposed several mechanisms for E6 activation of hTERT. In one model, Myc binds to E6 and translocates the E6/E6AP complex to the hTERT regulatory elements, as evidenced by chromatin immunoprecipitation (ChIP) data showing E6 and E6AP localized to the minimal hTERT promoter (45). In another model, the E6/E6AP complex stimulates the degradation of NFX-1, a constitutive repressor of hTERT transcription (17). A third group suggested that the USF transcription factor regulates hTERT transcription. These authors reported that Myc replaces USF at the hTERT promoter in cells expressing E6 (32). It has also been suggested that p300- and E6AP-dependent histone acetylation is involved in E6-induced telomerase activation (21).
While it has been reported that the activation of hTERT in human keratinocytes requires the binding of E6 to E6AP, only a few E6 mutations have been characterized (16, 30, 45). We had previously identified HPV16 E6 mutants that were defective in binding to E6AP yet produced immortal mammary epithelial cells (MECs), so we suspected that these mutants retained the ability to increase telomerase levels (31). We therefore sought to further characterize the relationship between E6AP association and telomerase activation by using an expanded series of E6 mutations.
MATERIALS AND METHODS
Retrovirus infection.
Transfection of LinX-A packaging cells and retrovirus production were performed as described elsewhere (41).
Cell culture.
Primary human foreskin keratinocytes were maintained in keratinocyte serum-free medium (KSFM; Invitrogen) or on feeder layers in F medium (14). Keratinocytes were infected with LXSN retroviruses expressing HPV16 E6 or one of its mutants and were selected with G418 (100 μg/ml) for 10 days. H1299 cells were maintained in RPMI-10% serum medium (Invitrogen).
Plasmids.
Flag-pcDNA3-16 E6 and its L37S mutant were kind gifts from Cheng-Ming Chiang (UT-Southwestern). Flag-pcDNA3-L110Q was generated by site-directed mutagenesis (QuikChange; Stratagene).
Real-time reverse transcription-PCR (RT-PCR).
Total RNA was isolated with an RNeasy kit (Qiagen). One microgram of total RNA was used for synthesis of first-strand cDNA with random primers and the Superscript III system (Invitrogen). Ten percent of the first-strand synthesis reaction product was used for TaqMan PCR on an MJ Research PCR cycler. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified using a SYBR green mix (S-7563; Molecular Probes) and was used as a loading control. The following primer pairs were used: GAPDH-G3S (ACCACAGTCCATGCCATCAC) and -G3A (TCCACCACCCTGTTGCTGTA) and hTERT forward (TTCTTGTTGGTGACACCTCACCTC) and reverse (ACAGGGAAGTTCACCACTGTCTTC) primers. The TaqMan hTERT probe (TGAGTATGGCTGCGTGGTGAACTT) was combined with the hTERT forward and reverse primers. E6 gene expression was quantified using a SYBR green mix with E6 forward (AAGCAACAGTTACTGCGACGTGAG) and reverse (CGGTCCACCGACCCTTATATT) primers. hTERT and E6 levels were normalized to GAPDH levels.
In vitro binding assay.
One microgram of glutathione S-transferase (GST)-E6AP protein was incubated with in vitro-translated 35S-labeled E6 proteins and protein A-Sepharose beads in low-salt association buffer (100 mM NaCl, 100 mM Tris-Cl [pH 8.0], 0.1% NP-40) for 1 h at 4°C (31). The beads were washed four times in the same buffer and loaded onto a 15% sodium dodecyl sulfate-acrylamide gel.
Quantitative binding assay.
In brief, E6AP and the human homologue of the Drosophila discs large tumor suppressor protein (hDLG) were fused in frame to the N terminus of bacterial alkaline phosphatase (BAP) and purified from Escherichia coli (2). E6 mutants were expressed as GST fusion proteins. Equivalent amounts of GST fusion proteins were immobilized on glutathione beads and plated into 96-well filter plates (500 ng/well). Saturation was determined by binding increasing amounts of E6AP-BAP or hDLG-BAP in 50 μl at room temperature for 1 h. Bound E6AP-BAP or hDLG-BAP was measured in relative light units using an Immunostar-AP substrate (Bio-Rad). Nonlinear regression was used to calculate disassociation constants (Kd) with Prism software.
ChIP assay.
Human foreskin keratinocytes infected with recombinant retroviruses expressing HPV16 E6 mutants were used for a ChIP assay, which was performed as described previously (50). Primers used to amplify endogenous hTERT have been described previously (45).
Coimmunoprecipitation experiments.
A total of 106 H1299 cells were transfected with 2 μg of Flag-pcDNA3 expressing HPV16 E6 or the L37S or L110Q mutant along with 2 μg of Flag-pcDNA3-E6AP. Cells were treated with the proteasome inhibitor MG132 and were harvested 24 h after transfection; cell pellets were frozen at −80°C, thawed on ice, and lysed with 50 mM Tris-Cl (pH 8.0), 1% NP-40, 150 mM NaCl, and 2 mM EDTA. E6AP-bound complexes were collected using an anti-E6AP antibody and were probed after immunoblotting with anti-Flag or anti-E6AP antibodies.
Antibodies.
The following antibodies were used in the ChIP assay: anti-HPV16 E6 (1), anti-Myc (9E10; Santa Cruz Biotechnology), anti-E6AP (N-14; Santa Cruz Biotechnology), and anti-actin (C-2; Santa Cruz Biotechnology). For immunoprecipitations and immunoblotting, we used anti-Flag antibody M2 (Sigma) and anti-E6AP antibody N-14 (Santa Cruz Biotechnology).
RESULTS AND DISCUSSION
We have previously reported the characterization of mutants of HPV16 E6 that could neither bind E6AP nor induce the degradation of p53 yet retained the ability to immortalize MECs (31). These observations superficially appeared to challenge the prevailing notion that both p53 pathway inactivation and reactivation of telomerase are necessary for the inhibition of replicative senescence in epithelial cells. Our subsequent investigations revealed that hAda3 is a critical effector of senescence signaling to p53 that involves its acetylation and stabilization and that E6 mutants capable of inducing hAda3 degradation block p53-dependent senescence (39; V. A. Shamanin, P. Sekaric, and E. J. Androphy, submitted for publication). In this study, we sought to understand whether and how these E6 mutants activate hTERT.
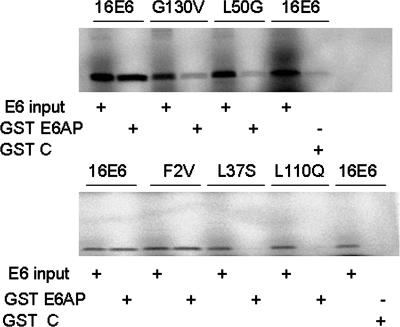
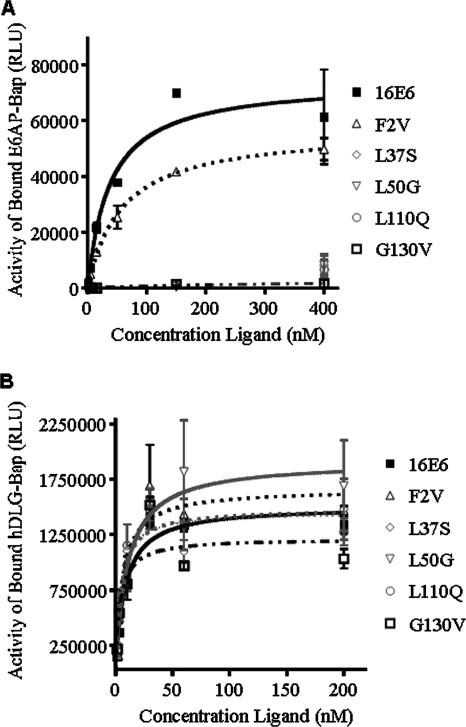
We selected five HPV16 E6 mutants for these investigations. The L37S and L110Q mutants were previously identified as defective in association with E6AP but competent at MEC immortalization (31). The L50G mutant has been reported not to bind E6AP (51) and did not immortalize MECs (Shamanin et al., submitted), and the G130V mutant had a phenotype identical to that of the L50G mutant in both these assays (Shamanin et al., submitted). The F2V mutant was included for comparison, since we previously reported that it bound E6AP and immortalized MECs (31). To correlate E6 binding to E6AP with hTERT induction, we initially evaluated the direct interaction of HPV16 E6 and the five mutants with E6AP by using a standard GST pulldown assay. As expected, wild-type HPV16 E6 and the F2V mutant efficiently bound to GST-E6AP, while the G130V, L50G, L37S, and L110Q mutants displayed no distinguishable binding above background (Fig. 1). However, this assay measures binding by using a fixed amount of GST-E6AP and undefined concentrations of wild-type and mutant E6 proteins. Therefore, we utilized a quantitative binding assay in which a fixed concentration of each GST-E6 protein was incubated with increasing concentrations of E6AP-BAP fusion protein (2; J. J. Cherry and E. J. Androphy, unpublished data). Only wild-type E6 and F2V E6 displayed E6AP-BAP binding that was significantly (P < 0.001 by two-way analysis of variance) greater than that for the GST control (Fig. 2); the Kd determined for wild-type HPV16 E6 and F2V E6 were 38 ± 15 nM and 57 ± 9 nM, respectively. The L50G, G130V, L37S, and L110Q E6 mutants were defective for binding even at the highest concentrations of the E6AP-BAP ligand (Fig. 2), so Kd could not be calculated. To verify that the GST-E6 fusion proteins were active, equal amounts were tested for binding to a BAP fusion containing the three PDZ domains of the hDLG protein (25). This domain remains intact in all the mutant GST-E6 fusion proteins studied here. As with the E6AP-BAP binding reactions, a constant amount of each GST-E6 fusion protein was incubated with increasing concentrations of the hDLG-BAP fusion protein (Fig. 2). All GST-E6 proteins bound to hDLG-BAP with similar efficiencies, at an average Kd of ∼6 nM and with no significant differences in the saturation binding curves (P > 0.05 by a two-way analysis of variance).
FIG. 1.
Mutational analysis of E6 binding to E6AP. GST-E6AP was incubated with in vitro-translated wild-type HPV16 E6 (16E6) and the G130V and L50G (top panel) and F2V, L37S, and L110Q (bottom panel) mutants. Background is shown in the last lane of each panel, in which GST alone (GST C) was used as a negative control.
FIG. 2.
Quantitative binding assay. Five hundred nanograms of each GST-E6 fusion protein was incubated with increasing concentrations of portions of E6AP or hDLG fused to BAP (ligand) in a final volume of 50 μl. After a wash, bound ligand was determined as relative light units (RLU). (A) Binding of GST-E6 proteins to increasing concentrations (0 to 400 nM) of E6AP-BAP. E6 and the F2V mutant bound E6AP and reached saturation; the L37S, L50G, L110Q, and G130V mutants did not bind to the ligand. (B) Binding of GST-E6 proteins to increasing concentrations (0 to 200 nM) of hDLG-BAP. All E6 proteins bound this ligand and reached saturation.
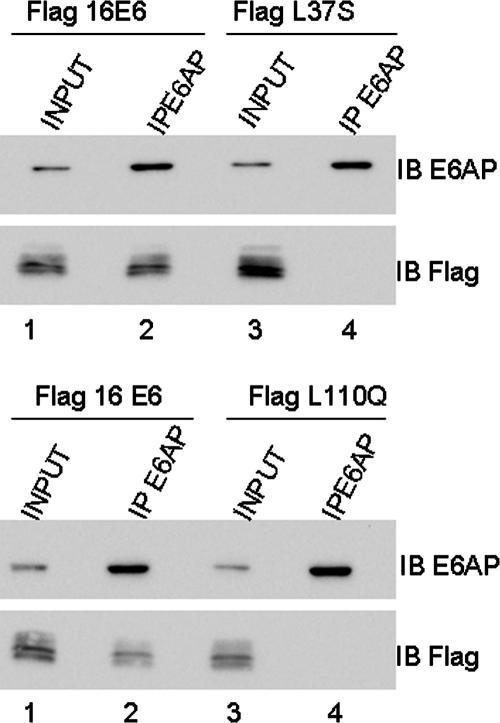
A recent report demonstrated association between E6 and E6AP by immunoprecipitation (5). To confirm our in vitro binding data, we used transient transfection of E6 and E6AP into H1299 cells, which do not express p53, so any interaction would be p53 independent. Because it has been reported that E6 binding induces autoubiquitination and destruction of E6AP, cells were treated with the proteasome inhibitor MG132 (5, 22). Immunoprecipitations with an anti-E6AP antibody showed that the complexes captured wild-type HPV16 E6 but not the L37S and L110Q mutants (Fig. 3), confirming the inability of these mutants to bind E6AP.
FIG. 3.
The HPV16 E6 L37S and L110Q mutants do not associate with E6AP in vivo. H1299 cells were transfected with Flag-pcDNA3-16 E6, -L37S, or -L110Q and Flag-pcDNA3-E6AP. Cells were treated with MG132 and harvested 24 h after transfection. To demonstrate the association between E6 and E6AP, cell lysates were immunoprecipitated with an anti-E6AP antibody, and bound complexes were immunoblotted with an anti-E6AP or anti-Flag antibody. E6AP immunoprecipitated only with wild-type E6, not with E6 L37S or L110Q.
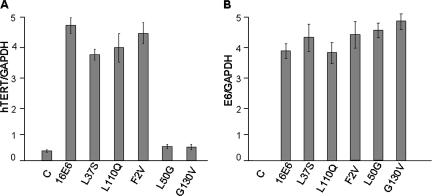
We then addressed the question of whether these E6AP binding-defective, MEC immortalization-competent E6 mutants could increase the expression of hTERT. For these experiments, we preferred to examine endogenous hTERT transcription rather than transfecting an hTERT promoter-based reporter construct, and human keratinocytes (HKs) were chosen, because they are the natural target cells of HPV. Recombinant retroviruses expressing wild-type E6 and E6 mutants were used to infect early-passage HKs, since these cells are inefficiently transfected using DNA. Wild-type and F2V mutant E6 proteins, which both bind E6AP, increased endogenous hTERT transcription in the infected HKs. The E6AP binding-defective G130V and L50G E6 mutants did not stimulate hTERT expression (Fig. 4A). However, the L37S and L110Q E6 mutants, which are equally defective for E6AP binding, activated endogenous hTERT expression to levels similar to those for wild-type E6 and the F2V mutant. Since hTERT induction might be dependent on E6 levels, we quantified E6 expression by real-time PCR in keratinocytes infected with these E6 mutants and showed that they have similar expression of E6 mRNA (Fig. 4B). Taken together, these results demonstrate that E6 binding to E6AP is not required for hTERT induction.
FIG. 4.
E6AP binding is not required for hTERT activation. Primary HKs were infected with HPV16 E6-expressing retroviruses. Real-time RT-PCR was performed on extracted RNA using TaqMan oligonucleotides specific for hTERT and SYBR green for E6 in parallel with GAPDH for normalization. (A) Ratio of hTERT to GAPDH amplification. The L37S and L110Q E6AP binding-defective mutants activated hTERT to levels similar to those for wild-type HPV16 E6 and the F2V mutant. The L50G and G130V mutants did not increase hTERT transcript levels more than the control (C) retrovirus. (B) Ratio of E6 expression to GAPDH expression.
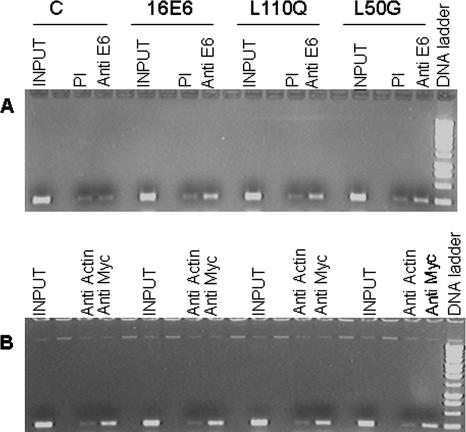
Another aspect of the proposed model is that HPV16 E6 is present at the hTERT promoter (45). We performed ChIP experiments to test for wild-type and mutant HPV16 E6 proteins on the endogenous hTERT promoter in keratinocytes following retrovirus infections. An anti-E6 peptide serum, but not preimmune serum, immunoprecipitated the hTERT promoter DNA, as predicted (Fig. 5A). We then queried whether the E6AP binding-defective E6 mutants could still localize to the hTERT promoter. We selected for comparison two E6AP binding-defective E6 mutants: the L110Q mutant, which induced hTERT, and the L50G mutant, which did not. Notably, both the L110Q and L50G E6 proteins associated with the endogenous hTERT promoter by ChIP assay (Fig. 5A), implying that E6AP binding is not necessary to target E6 to the hTERT promoter. Interestingly, the L50G data suggest that E6 localization at the hTERT promoter is not sufficient for hTERT induction, while the L37S and L110Q results suggest the involvement of an activity distinct from E6AP binding.
FIG. 5.
ChIP assays for E6 and Myc at the hTERT promoter. HKs were infected with HPV16 E6-expressing retroviruses and treated with formaldehyde. Extracts were immunoprecipitated, the cross-linking reversed, and PCR performed with primers to amplify hTERT DNA. The input is the extract prior to immunoprecipitation that was subject to PCR. (A) ChIP assay with antiserum to E6 or preimmune serum (PI). E6AP binding-defective HPV16 E6 mutants associate with the endogenous hTERT promoter. E6 mutants were detected at the hTERT promoter by using an anti-E6 peptide antibody. No E6 signal was detected in cells expressing retroviral backbone DNA (lanes C). (B) A ChIP assay using an anti-Myc antibody detects Myc at the hTERT promoter in HKs infected with control (lanes C) and E6-expressing retroviruses. An anti-actin antibody was used as a control.
E6AP has been detected at the hTERT promoter in the presence of E6, suggesting that the E6/E6AP complex might target a telomerase repressor for ubiquitin-mediated proteolysis (30). We performed ChIP assays using a commercially available anti-E6AP antibody (N-14; Santa Cruz) and an anti-E6AP peptide antiserum, both confirmed to recognize cotransfected epitope tagged E6AP by immunoprecipitation (data not shown). These ChIP experiments were unable to detect E6AP on the endogenous hTERT promoter in HKs in the presence or absence of wild-type E6 (data not shown).
Binding of Myc to specific E boxes within the hTERT enhancer elements activates hTERT transcription (9, 19). Myc and its associated E box sites are also critical for E6 induction of hTERT transcription, and experimental evidence indicates that Myc binds and transfers E6 to these regulatory sites (16, 32, 35, 45). We therefore tested for the presence of Myc at the hTERT promoter and asked whether this correlates with E6-mediated activation. We performed ChIP assays using extracts from HKs infected with a retrovirus expressing either wild-type E6 or the L110Q (hTERT-activating) or L50G (defective for hTERT induction) E6AP binding-defective mutant. Binding of Myc to the hTERT promoter was detected in HKs in the absence of E6 as well as in cells expressing wild-type or mutant E6 (Fig. 5B), while no hTERT PCR products were visualized by using a control anti-actin antibody. These results reproduce prior data (30, 45) showing that Myc bound to the hTERT promoter in the absence of E6. In contrast, a recent study detected Myc at the hTERT promoter only in cells expressing E6 and found that E6 increased Myc protein levels (32). In our experiments, E6 did not alter the levels of total Myc protein in the HKs (data not shown), in agreement with other reports (16, 35, 45).
The discrepancies in Myc protein levels may be explained by differences in cell culture methods. In our experiments, HKs were maintained on feeder layers before retrovirus infection, with subsequent transfer to KSFM. It has been shown that keratinocyte growth conditions (feeder layers versus a minimal medium such as KSFM) can modulate telomerase expression (14). Additionally, Myc is known to interact with HAT complexes SAGA and GCN5 (13, 29), and GCN5, TIP60, CBP, and p300 have been shown to mediate Myc acetylation (11, 37, 46). Ablation of p300 also increased histone acetylation and caused hTERT activation, implying that p300 acts as a repressor of telomerase (21). E6 has been reported to interact with HAT complex proteins such as p300 and hAda3 (27, 36) and to inhibit p53 acetylation (39, 43). It is therefore possible that acetylation of Myc, rather than its phosphorylation or alterations in the total levels of Myc or its binding partners Mad and Max, is involved in hTERT activation (45). Future studies should reveal whether E6 alters Myc acetylation and whether this regulates hTERT activation.
Our data appear to conflict with the concept that E6 must bind to E6AP in order to activate hTERT (16, 30). Using reporter and telomere repeat amplification protocol assays, previous studies showed that the HPV16 8S9A10T E6 mutant, which binds E6AP, could activate telomerase while the E6AP binding-defective Δ9-13 mutant, with a 5-amino-acid deletion, was inactive. Another study suggested that E6AP binding is important for telomerase activation, since E6AP binding-defective deletion mutants (Δ123-127, Δ118-122, and Δ146-151) of E6 did not induce telomerase in a telomere repeat amplification protocol assay (24, 26). We investigated the activation of endogenous hTERT by real-time RT-PCR. By analyzing E6 mutants with single amino acid substitutions, we find that E6/E6AP interaction is not required for the activation of the endogenous hTERT promoter. Interestingly, the leucine at position 110, which was able to stimulate hTERT and induce MEC immortalization (31), has been reported to be buried at a hydrophobic interface of the proposed E6 structural model (33). Substitution of charged glutamine is likely to disrupt this fold. However, the reported structure was modeled from nuclear magnetic resonance data for a C-terminal portion of E6 and may not accurately reproduce the native conformation of the E6 protein.
The E6/E6AP complex was reported to bind two isoforms of NFX1: NFX1-91 and NFX1-123 (17). NFX1 was demonstrated to repress hTERT transcription, and E6 interfered with this repression by destabilizing NFX1-91, presumably through E6AP-mediated ubiquitination. Our data demonstrate that E6AP is not required for hTERT activation and therefore suggest that NFX1-91 destabilization is not necessary or that another E6-dependent pathway may specifically target NFX1-91. This is consistent with recent observations that E6-induced p53 degradation in vivo may occur through E6AP-dependent and E6AP-independent mechanisms (6, 7, 40).
The NFX1-123 protein was a strong activator of hTERT in human keratinocytes expressing HPV16 E6 (23). E6 bound NFX1-123 in the absence of E6AP and did not induce its degradation. These authors suggested that E6 recruits NFX1-123 to the hTERT promoter. Since we show that the E6AP binding-defective L110Q and L37S E6 mutants activate hTERT, one possible scenario is that these mutants retain the ability to interact with NFX-123 and to target it to the hTERT promoter. The L50G mutant does not bind E6AP but was found localized to the hTERT promoter; still, it did not induce hTERT transcription. It is possible that the L50G mutant lacks the ability to interact with NFX-123.
Our data reveal a direct correlation between telomerase activation and immortalization, since mutants that induce hTERT (the F2V, L37S, and L110Q mutants [Fig. 4A]) immortalize MECs (31). The Δ9-13, Δ123-127, and Δ146-151 E6 deletion mutants were defective for telomerase activation and failed to immortalize MECs (24). Taken together, our results strongly suggest that induction of hTERT is necessary for MEC immortalization. We have not yet studied the E6 activities necessary for human keratinocyte immortalization due to the confounding requirement for E7. The characterization of these HPV16 E6 mutants should facilitate such studies.
In conclusion, we demonstrate that E6 binding to the ubiquitin ligase E6AP is not necessary for E6 localization to the hTERT promoter or for induction of hTERT transcription. Moreover, recruitment of E6 to the hTERT promoter is not sufficient for telomerase activation, since the E6 L50G mutant was present at the hTERT promoter but did not activate hTERT. Further studies are required to determine the precise mechanism by which E6 stimulates hTERT transcription.
Acknowledgments
Flag-pcDNA3-16 E6 and the L37S mutant were kind gifts from Cheng-Ming Chiang (UT Southwestern). We thank Vladimir A. Shamanin for critical reading of the manuscript and Christina Gutierrez for technical assistance.
This work was supported by NIH grant RO1 CA107394.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Androphy, E. J., N. L. Hubbert, J. T. Schiller, and D. R. Lowy. 1987. Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J. 6989-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baleja, J. D., J. J. Cherry, Z. Liu, H. Gao, M. C. Nicklaus, J. H. Voigt, J. J. Chen, and E. J. Androphy. 2006. Identification of inhibitors to papillomavirus type 16 E6 protein based on three-dimensional structures of interacting proteins. Antivir. Res. 7249-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwood, E. M., and R. N. Eisenman. 1991. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 2511211-1217. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279349-352. [DOI] [PubMed] [Google Scholar]
- 5.Brimer, N., C. Lyons, and S. B. Vande Pol. 2007. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology 358303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camus, S., M. Higgins, D. P. Lane, and S. Lain. 2003. Differences in the ubiquitination of p53 by Mdm2 and the HPV protein E6. FEBS Lett. 536220-224. [DOI] [PubMed] [Google Scholar]
- 7.Camus, S., S. Menendez, C. F. Cheok, L. F. Stevenson, S. Lain, and D. P. Lane. 2007. Ubiquitin-independent degradation of p53 mediated by high-risk human papillomavirus protein E6. Oncogene 264059-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269529-531. [DOI] [PubMed] [Google Scholar]
- 9.Cong, Y. S., J. Wen, and S. Bacchetti. 1999. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet. 8137-142. [DOI] [PubMed] [Google Scholar]
- 10.Dimri, G. P., J. L. Martinez, J. J. Jacobs, P. Keblusek, K. Itahana, M. Van Lohuizen, J. Campisi, D. E. Wazer, and V. Band. 2002. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 624736-4745. [PubMed] [Google Scholar]
- 11.Faiola, F., X. Liu, S. Lo, S. Pan, K. Zhang, E. Lymar, A. Farina, and E. Martinez. 2005. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol. Cell. Biol. 2510220-10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, E. J., and S. C. Wright. 2001. S-phase-specific expression of the Mad3 gene in proliferating and differentiating cells. Biochem. J. 359361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, B., J. Quintero, and C. C. Baker. 2003. Keratinocyte growth conditions modulate telomerase expression, senescence, and immortalization by human papillomavirus type 16 E6 and E7 oncogenes. Cancer Res. 637815-7824. [PubMed] [Google Scholar]
- 15.Gao, Q., L. Singh, A. Kumar, S. Srinivasan, D. E. Wazer, and V. Band. 2001. Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells. J. Virol. 754459-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 757198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 182269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goueli, B. S., and R. Janknecht. 2003. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene 228042-8047. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg, R. A., R. C. O'Hagan, H. Deng, Q. Xiao, S. R. Hann, R. R. Adams, S. Lichtsteiner, L. Chin, G. B. Morin, and R. A. DePinho. 1999. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 181219-1226. [DOI] [PubMed] [Google Scholar]
- 20.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 134918-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James, M. A., J. H. Lee, and A. J. Klingelhutz. 2006. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 1191878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, W. H., S. L. Beaudenon, A. L. Talis, J. M. Huibregtse, and P. M. Howley. 2000. Human papillomavirus type 16 E6 induces self-ubiquitination of the E6AP ubiquitin-protein ligase. J. Virol. 746408-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzenellenbogen, R. A., E. M. Egelkrout, P. Vliet-Gregg, L. C. Gewin, P. R. Gafken, and D. A. Galloway. 2007. NFX1-123 and poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16 E6-expressing cells. J. Virol. 813786-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 39684-88. [DOI] [PubMed] [Google Scholar]
- 25.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 9411612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 38079-82. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, A., Y. Zhao, G. Meng, M. Zeng, S. Srinivasan, L. M. Delmolino, Q. Gao, G. Dimri, G. F. Weber, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 225801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, S. Y., and S. J. Elledge. 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113881-889. [DOI] [PubMed] [Google Scholar]
- 29.Liu, X., J. Tesfai, Y. A. Evrard, S. Y. Dent, and E. Martinez. 2003. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J. Biol. Chem. 27820405-20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, X., H. Yuan, B. Fu, G. L. Disbrow, T. Apolinario, V. Tomaic, M. L. Kelley, C. C. Baker, J. Huibregtse, and R. Schlegel. 2005. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 28010807-10816. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 737297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMurray, H. R., and D. J. McCance. 2003. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J. Virol. 779852-9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nominé, Y., M. Masson, S. Charbonnier, K. Zanier, T. Ristriani, F. Deryckere, A. P. Sibler, D. Desplancq, R. A. Atkinson, E. Weiss, G. Orfanoudakis, B. Kieffer, and G. Trave. 2006. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. Cell 21665-678. [DOI] [PubMed] [Google Scholar]
- 34.Oh, S., Y. H. Song, J. Yim, and T. K. Kim. 2000. Identification of Mad as a repressor of the human telomerase (hTERT) gene. Oncogene 191485-1490. [DOI] [PubMed] [Google Scholar]
- 35.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 755559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 185061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, J. H., Y. Du, P. G. Ard, C. Phillips, B. Carella, C. J. Chen, C. Rakowski, C. Chatterjee, P. M. Lieberman, W. S. Lane, G. A. Blobel, and S. B. McMahon. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 2410826-10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 631129-1136. [DOI] [PubMed] [Google Scholar]
- 39.Sekaric, P., V. A. Shamanin, J. Luo, and E. J. Androphy. 2007. hAda3 regulates p14ARF-induced p53 acetylation and senescence. Oncogene 266261-6268. [DOI] [PubMed] [Google Scholar]
- 40.Shai, A., M. L. Nguyen, J. Wagstaff, Y. H. Jiang, and P. F. Lambert. 2007. HPV16 E6 confers p53-dependent and p53-independent phenotypes in the epidermis of mice deficient for E6AP. Oncogene 263321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shamanin, V. A., and E. J. Androphy. 2004. Immortalization of human mammary epithelial cells is associated with inactivation of the p14ARF-p53 pathway. Mol. Cell. Biol. 242144-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takakura, M., S. Kyo, T. Kanaya, H. Hirano, J. Takeda, M. Yutsudo, and M. Inoue. 1999. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 59551-557. [PubMed] [Google Scholar]
- 43.Thomas, M. C., and C. M. Chiang. 2005. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell 17251-264. [DOI] [PubMed] [Google Scholar]
- 44.Vaziri, H., and S. Benchimol. 1998. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 8279-282. [DOI] [PubMed] [Google Scholar]
- 45.Veldman, T., X. Liu, H. Yuan, and R. Schlegel. 2003. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 1008211-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vervoorts, J., J. M. Luscher-Firzlaff, S. Rottmann, R. Lilischkis, G. Walsemann, K. Dohmann, M. Austen, and B. Luscher. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 4484-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, J., L. Y. Xie, S. Allan, D. Beach, and G. J. Hannon. 1998. Myc activates telomerase. Genes Dev. 121769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, D., N. Popov, M. Hou, Q. Wang, M. Bjorkholm, A. Gruber, A. R. Menkel, and M. Henriksson. 2001. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 983826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, G. L., J. D. Bradley, L. D. Attardi, and E. H. Blackburn. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344126-132. [DOI] [PubMed] [Google Scholar]
- 50.Yu, T., Y. C. Peng, and E. J. Androphy. 2007. Mitotic kinesin-like protein 2 binds and colocalizes with papillomavirus E2 during mitosis. J. Virol. 811736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmermann, H., R. Degenkolbe, H. U. Bernard, and M. J. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 736209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]