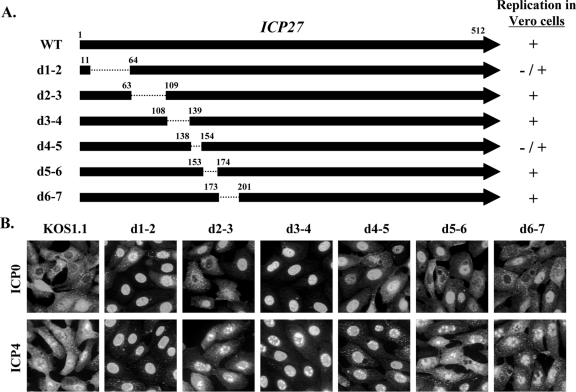
FIG. 1.
Effect of N-terminal ICP27 deletions on the nucleocytoplasmic distribution of ICP0 and ICP4 during viral infection. (A) Diagram of the ICP27 polypeptides produced by the viral deletion mutants used in this study. The top arrow represents the 512-residue ICP27 polypeptide, and the arrows below represent deleted proteins encoded by the various viral mutants. The residues bordering the deleted regions (dotted lines) are indicated. The ability of the mutants to replicate in Vero cells (26) is shown at right. (B) ICP0 and ICP4 localization. Vero cells were infected at an MOI of 10 with WT HSV-1 or the mutant viruses as shown. Cells were fixed at 6 hpi and processed for ICP0 and ICP4 immunofluorescence.