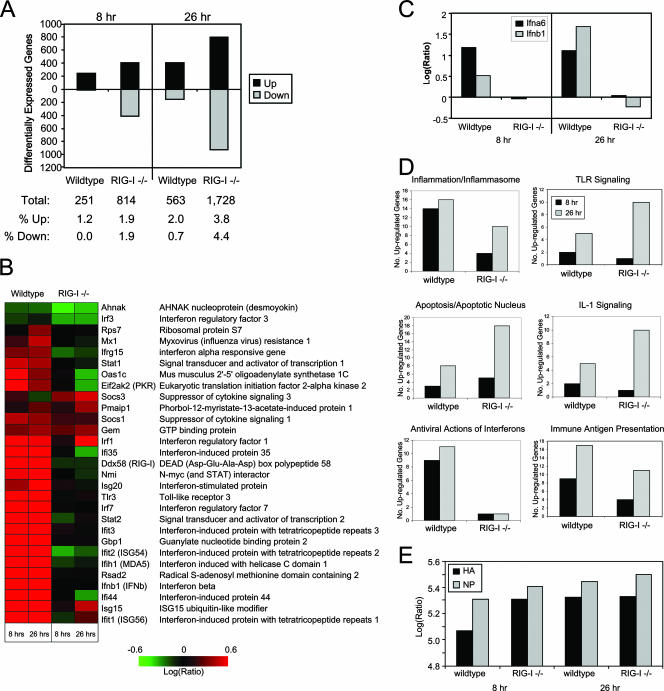
FIG. 5.
Identification of RIG-I-responsive genes by functional genomics analyses. wt or RIG-I−/− mouse fibroblasts were either mock infected or infected with influenza virus A/PR/8/34 at a multiplicity of infection of 5. At 8 and 26 h postinfection, cells were collected and cellular RNA extracted for DNA microarray or qPCR analyses as described in the text. (A) (Top) Numbers of differentially expressed genes (up- or down-regulated more than twofold) as identified by DNA microarray analyses in wt and RIG-I−/− cells following influenza virus A/PR/8/34 infection at 8 and 26 h relative to expression in mock-infected cells. (Bottom) Table showing the total numbers of differentially expressed genes and the breakdown of genes that were differentially expressed as a percentage of total genes analyzed. (B) Heat map showing the differential expression of a bioset of RIG-I-responsive genes following influenza virus A/PR/8/34 infection. (C) Gene expression of IFN-α6 and IFN-β was verified by qualitative real-time PCR. (D) Numbers of genes, segregated by their known functions, whose expression was determined by a DNA microarray to be induced more than twofold following influenza virus A/PR/8/34 infection for 8 and 26 h. Additional data are available at http://expression.viromics.washington.edu. (E) Qualitative real-time PCR analysis for viral HA protein and NP in RNA extracted from wt or RIG-I−/− cells that were either mock infected or infected with influenza virus A/PR/8/34 for 8 or 26 h.