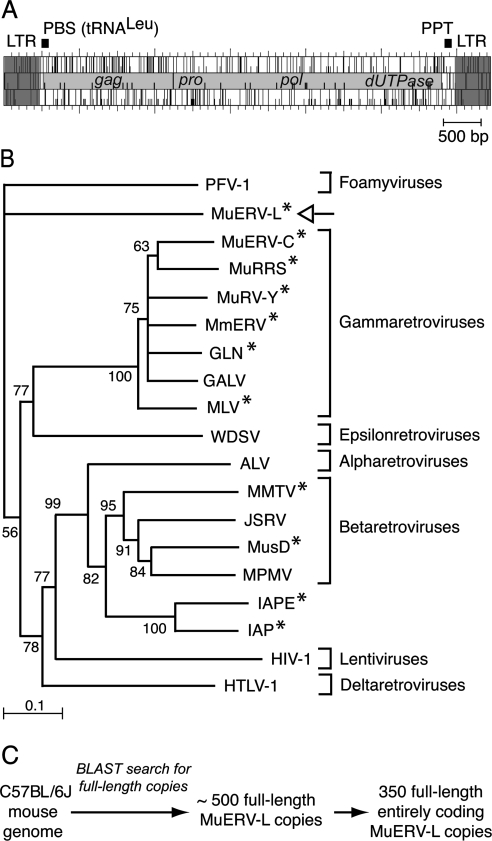
FIG. 1.
Structure and phylogeny of MuERV-L elements. (A) Genomic organization of an MuERV-L provirus, with the LTRs (in dark gray) flanking two ORFs homologous to the retroviral gag and pol genes (in light gray). The pol gene further discloses domains homologous to retroviral pro and dUTPase genes. A primer binding site (PBS) complementary to tRNALeu and a polypurine track (PPT) can be identified. (B) Phylogeny of retroviruses, based on their reverse transcriptase domain. The tree was constructed by the neighbor-joining method using the seven blocks of conserved residues found in the reverse transcriptase domain of all retroelements and was rooted with non-LTR retrotransposons. All sequences are readily accessible from GenBank and previous reports (e.g., reference 15). Percent bootstrap values obtained from 1,000 replicates are indicated. The retroviruses “endogenized” in the mouse genome are marked (*). The seven retroviral genera are indicated on the right. PFV, primate foamy virus; MuRRS, murine retrovirus-related DNA sequence; MuRV-Y, murine retrovirus Y associated; MmERV, Mus musculus ERV; GLN, murine retrovirus using tRNAGln; GALV, gibbon ape leukemia virus; MLV, murine leukemia virus; WDSV, walleye dermal sarcoma virus; ALV, avian leukosis virus; MMTV, mouse mammary tumor virus; JSRV, jaagsiekte sheep retrovirus; MusD, Mus musculus type D retrovirus; MPMV, Mason-Pfizer monkey virus; IAPE, IAP with an envelope gene; HIV-1, human immunodeficiency virus type 1; HTLV-1, human T-cell leukemia virus type 1. (C) Search for full-length, entirely coding MuERV-L copies in the C57BL/6J genome. Blast search was carried out with the NCBI m36 mouse assembly (April 2006 release) and yielded 489 full-length MuERV-L copies among which 350 contain the two complete gag and pol ORFs.