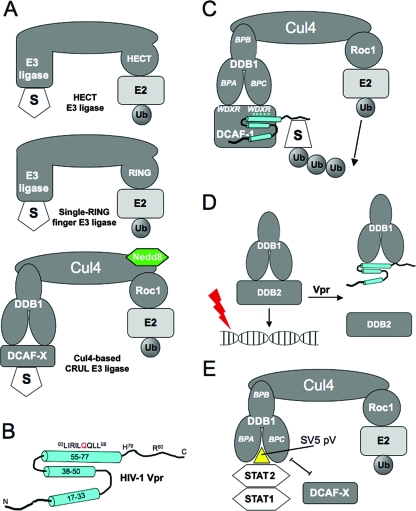
FIG. 1.
E3 ubiquitin ligases and their interactions with viral proteins. (A) The three main types of E3 ligases. (B) The nuclear magnetic resonance structure of HIV-1 Vpr. Cylinders represent α-helices, and lines are unstructured regions. (C) The Vpr-UPS model predicts the interaction of Vpr with DCAF1 and a putative degradation substrate, “S.” BPA, BPB, and BPC denote the three beta-propeller domains of DDB1. Le Rouzic et al. (34) found that the WDXR motifs of DCAF1 are required for the interaction with Vpr. For simplicity, we have represented Vpr interacting with the WDXR motif on BPC; there is no evidence to support which of the two WDXR motifs is required for Vpr interaction. (D) The Vpr-UV-DDB model predicts that the interaction of Vpr with DDB1 displaces DDB2 and prevents the DDB1-DDB2 complex, also known as UV-DDB, from recognizing damaged DNA. (E) SV5 protein V (pV) interacts with DDB1 and displaces DCAF, creating a new interface that recruits STAT2 as an adaptor, which then recruits STAT1 for ubiquitination. Ub, ubiquitin.