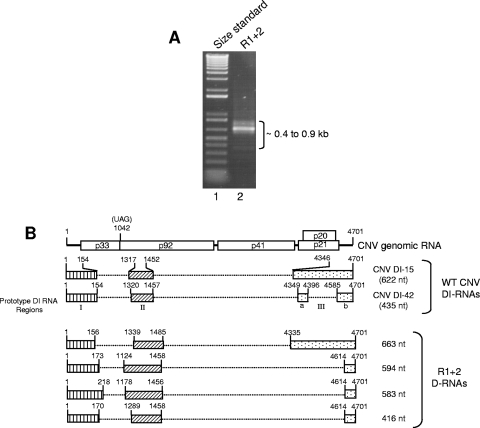
FIG. 4.
Characterization of CNV DI-like RNAs in T=1 particles of mutant R1+2. (A) EtBr-stained agarose gel showing RT-PCR products of RNA purified from T=1 particles of R1+2 (lane 2). RT-PCR primers corresponded to the 5′ and 3′ termini of CNV gRNA. Lane 1 contains a molecular size standard (1-kb Plus DNA ladder; Invitrogen). The approximate sizes of the RT-PCR products are indicated. (B) Structure of R1+2 DI-like RNAs. The structure of CNV genome RNA is shown at the top along with the structures of two previously described prototype CNV DI RNAs (11). The three major regions of the CNV genome that are retained in the prototype DI RNAs (I, II, and III) are depicted by the boxed regions in CNV DI-15 and DI-42. Region III of DI-42 is divided into subregions IIIa and IIIb as previously described (11). The bottom shows structures of DI-like RNAs deduced from nucleotide sequence analysis. The shading patterns used correspond to those of the prototype DI-like RNAs. Numbers at the 5′ and 3′ termini of the boxes indicate the nucleotide positions at the boundaries of the box region relative to CNV gRNA. The size in nucleotides of each DI or DI-like RNA is shown on the right. D-RNA, defective RNA.