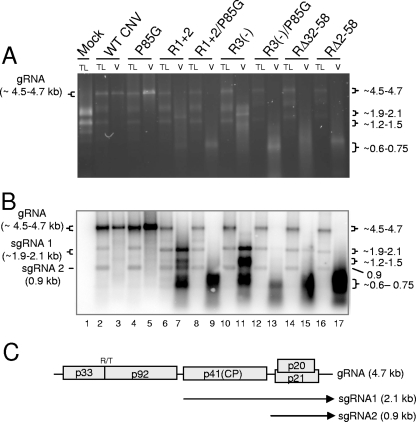
FIG. 7.
Agarose gel (A) and Northern blot analysis (B) of total leaf RNA extracts and virion RNAs of the indicated mutants. Leaves infected with each of the mutants were ground in liquid nitrogen. Total leaf RNA was extracted from one portion of the frozen mixture, and virions were extracted from another portion. Equal amounts of total leaf RNA (TL) or RNA extracted from virions (V) were loaded onto a denaturing agarose gel and stained with EtBr. A duplicate gel was blotted and hybridized with a mixture of two probes, one corresponding to the 5′ terminal 447 nt and the other to the 3′ terminal 428 nt of CNV gRNA. (C) Genome structure of WT CNV RNA showing the sizes and origins of sgRNA 1 (2.1 kb) and sgRNA 2 (0.9 kb). The values on the left in panel A correspond to the known size of gRNA in WT CNV virions and total leaf RNA extracts (4.7 kb) as well as the range of sizes predicted for the gRNA of the various mutants (summarized in Table 1). The values on the right in panel A correspond to the approximate sizes of the RNA species detected in particles of the various mutants (Table 1). The values on the left in panel B correspond to the known and predicted sizes of gRNA and sgRNA1 and -2 that are detected in blots of WT and mutant-infected total leaf RNA extracts. The range of sizes shown for gRNA and sgRNA2 reflects the sizes of the various mutant constructs (Table 1). The size of the sgRNA2 is not affected in the mutants. The values on the right in panel B correspond to the approximate sizes of various species detected in blotted total leaf and virion RNA extracts (Table 1). Sizes were estimated using the Invitrogen 0.24- to 9.5-kb RNA ladder as well as the known sizes of CNV gRNA, sgRNA1, and sgRNA2.