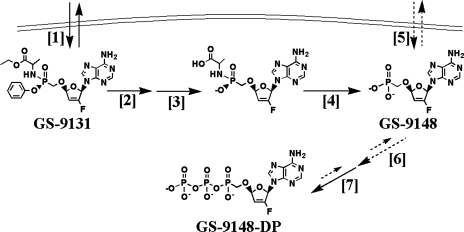
FIG. 7.
Putative mechanism of intercellular activation of GS-9131 and GS-9148. Arrows with solid lines represent efficient metabolic steps, while arrows with dashed lines represent more inefficient metabolic processes. First, the lipophilic prodrug GS-9131 freely permeates the cell [1] and is cleaved by lysosomal protease cathepsin A and possibly by other cellular hydrolases [2]. Phenol is lost by spontaneous nucleophilic release [3], and l-alanine is then released through hydrolysis of the P-N bond yielding GS-9148, either by a previously described phosphoamidase (29) or spontaneously due to the low pH present in the lysosomal compartment [4]. The poor cell membrane permeation of the nucleotide analog GS-9148 [5] is then phosphorylated by the rate-limiting first phosphorylation step, likely catalyzed by an adenylate kinase [6], followed by the more efficient second phosphorylation step, putatively catalyzed by creatine kinase or other kinases [7], generating the active metabolite GS-9148-DP.