Crystal structure of the ligand-binding domain of androgen receptor in complex with LGD2226.
Keywords: selective modulators, LGD2226, androgen receptor ligand-binding domain, SARM
Abstract
The androgen receptor (AR) is a ligand-inducible steroid hormone receptor that mediates androgen action, determining male sexual phenotypes and promoting spermatogenesis. As the androgens play a dominant role in male sexual development and function, steroidal androgen agonists have been used clinically for some years. However, there is a risk of potential side effects and most steroidal androgens cannot be dosed orally, which limits the use of these substances. 1,2-Dihydro-6-N,N-bis(2,2,2-trifluoroethyl)amino-4-trifluoromethyl-2-quinolinone (LGD2226) is a synthetic nonsteroidal ligand and a novel selective AR modulator. The crystal structure of the complex of LGD2226 with the androgen receptor ligand-binding domain (AR LBD) at 2.1 Å was solved and compared with the structure of the AR LBD–R1881 complex. It is hoped that this will aid in further explaining the selectivity of LGD2226 observed in in vitro and in vivo assays and in developing more selective and effective therapeutic agents.
1. Introduction
The androgen receptor (AR) is a ligand-inducible steroid hormone receptor that is widely distributed throughout the body and is involved in diverse activities, but its primary and dominant functions are in male sex development and differentiation (Collins et al., 2003 ▶; Gottlieb et al., 2005 ▶; Heinlein & Chang, 2002 ▶; Pelletier, 2000 ▶). It is a member of the nuclear receptor superfamily, with which it shares structural and functional similarity. It contains three principal domains, (i) a hypervariable N-terminal domain which regulates transcriptional activity, (ii) a central highly conserved DNA-binding domain and (iii) a large C-terminal ligand-binding domain (AR LBD), and a short linker between the DNA-binding domain and the AR LBD (Gao et al., 2005 ▶). AR is the chief regulatory intracellular transcription factor for genes involved in the proliferation and differentiation of the prostate. Androgen deprivation has been the standard therapy for advanced and metastatic prostate cancer for over half a century.
The function of AR is regulated by the binding of androgens, which initiates sequential conformational changes of the receptor that affect receptor–protein interactions and receptor–DNA interactions. Potential uses of androgens include male hormone-replacement therapy, male contraception and treatment of bone disorders, wasting diseases and female androgen deficiency, amongst many others. Theoretically, androgen therapy could be almost as widely used as female sex-hormone therapies, but this would require more selective and effective therapeutic agents.
The androgens can be classified as steroidal and nonsteroidal based on their structures. The endogenous androgens and modified steroidal ligands have some shortcomings that limit their general use, mainly their potential for side effects and their mode of administration. In contrast to the agonists, nonsteroidal androgen antagonists have been marketed for many years (Labrie, 1993 ▶; Hamann et al., 1998 ▶).
LGD2226 is a member of a novel class of orally active selective androgen receptor modulators (SARMs) that have the potential to provide improved therapeutic benefits while reducing the risk of side effects. In animal models it prevents bone loss, stimulates the formation of new bone and stimulates muscle growth, while having reduced stimulatory effects on the prostate (Rosen & Negro-Vilar, 2002 ▶; van Oeveren et al., 2006 ▶). To gain further insight into the structure–activity relationships of androgens and to determine whether the observed tissue selectivity has a direct basis on a molecular level, we solved the crystal structure of human AR LBD in complex with LGD2226. A comparison of this structure with the human AR LBD structure with the ligand metribolone (R1881; Matias et al., 2000 ▶) provides clues to achieving tissue selectivity and obtaining more effective potential therapeutic agents.
2. Materials and methods
The LBD of human AR (amino-acid residues 663–919) was amplified by polymerase chain reaction technology using the appropriate primers and inserted between the XbaI site and SalI site of a pGEX-KG vector (ATCC). The resulting fusion proteins consisted of a glutathione S-transferase, containing a carboxy-terminal thrombin cleavage site, optimized by a glycine-rich ‘kinker’ region followed by the corresponding human AR LBD. The constructs were then transformed into Escherichia coli strain BL21 (DE3). Expression took place as described by Matias et al. (2000 ▶) and was carried out in 2×YT medium in the presence of ampicillin (200 µg ml−1) supplemented with 10 µM LGD2226. Expression was induced with 40 µM isopropyl β-d-thiogalactoside (IPTG) at 288 K for about 16 h. The fused protein was purified by affinity chromatography using a glutathione-Sepharose column (GST column). The eluted protein was diluted immediately with buffer consisting of 0.1 M HEPES pH 7.2, 0.15 M NaCl, 0.5 mM EDTA, 10%(v/v) glycerol, 1 mM DTT, 10 µM LGD2226 and 0.1%(w/v) n-octyl-β-glucoside (buffer I) to about 1 mg ml−1. Cleavage with 1 unit of thrombin per millilitre was performed overnight at 277 K and the AR LBD complex (which, after thrombin treatment, produces an LBD consisting of residues 663–919 with a tag remnant of sequence GSPGNFRWWWWNSR attached to the N-terminus) was eluted using buffer I, while the GST column was recovered with 15 mM reduced glutathione in buffer I to elute the GST tag. The AR LBD complex was further purified using a Resource S (GE Biosciences) column before crystallization.
For crystallization, similar conditions were used as for the AR LBD–R1881 complex crystals, using the sitting-drop vapour-diffusion method, by mixing 2 µl of protein solution (5 mg ml−1 in buffer I) with an equal volume of the reservoir solution, which contained 0.3–0.4 M Na2HPO4/K2HPO4, 5%(w/v) PEG 400, 0.1 M (NH4)2HPO4, 0.1 M Tris pH 8.9, and equilibrating against 1 ml reservoir solution. Single crystals grew to suitable dimensions in 2–4 d. Crystals were cryoprotected in Paratone-N oil (Hampton Research) and frozen at 79 K, and X-ray data were collected using an FR-E diffraction system at the Institute of Biophysics (Beijing, People’s Republic of China). The data were processed using the CrystalClear software package (Rigaku).
We carried out molecular replacement using MOLREP (Vagin & Teplyakov, 1997 ▶) from CCP4 (Collaborative Computational Project, Number 4, 1994 ▶) with the coordinates of the AR LBD–R1881 complex (PDB code 1e3g; the solvent molecules and R1881 were removed) as the initial model. Refinement was carried out using the programs REFMAC (Murshudov et al., 1997 ▶) and CNS (Brünger et al., 1998 ▶). A sample containing a random 5% of the total reflections in the data set was excluded for R free calculations. After rigid-body refinement, electron density for the LGD2226 ligand was clearly visible in 2F o − F c and F o − F c maps. A model for the ligand was constructed using O (Jones et al., 1991 ▶) and CNS topology and parameter dictionaries were built using XPLO2D (Kleywegt & Jones, 1998 ▶). In the final refinement at 2.1 Å, the crystallographic R factor and R free were 22.1 and 25.3%, respectively, with good stereochemistry. Statistics of the data collection and final structure are summarized in Table 1 ▶. Figures were produced using PyMOL (http://www.pymol.org) and LIGPLOT (Wallace et al., 1995 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 56.64, b = 66.02, c = 73.98, α = 90.00, β = 90.00, γ = 90.00 |
| Resolution range | 28.35–2.10 (2.18–2.10) |
| Total No. of reflections | 197184 |
| No. of unique reflections | 16570 |
| Average redundancy | 11.9 (3.92) |
| Completeness (%) | 99.5 (99.2) |
| Rmerge† | 0.055 (0.283) |
| Reduced χ2 | 0.55 (0.59) |
| Output 〈I/σ(I)〉 | 23.5 (5.5) |
| R factor‡ (%) | 22.1 |
| Rfree‡ (%) | 25.3 |
| R.m.s. deviation from ideal geometry | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.46 |
R
merge = 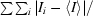
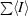 , where 〈I〉 is the mean intensity of N reflections with intensities Ii and common indices h, k and l.
, where 〈I〉 is the mean intensity of N reflections with intensities Ii and common indices h, k and l.
R factor = 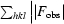 −
− 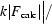
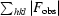 , where F
obs and F
calc are the observed and calculated structure factors, respectively. For R
free, the sum extends over a subset of reflections (5%) excluded from all stages of refinement.
, where F
obs and F
calc are the observed and calculated structure factors, respectively. For R
free, the sum extends over a subset of reflections (5%) excluded from all stages of refinement.
3. Results and discussion
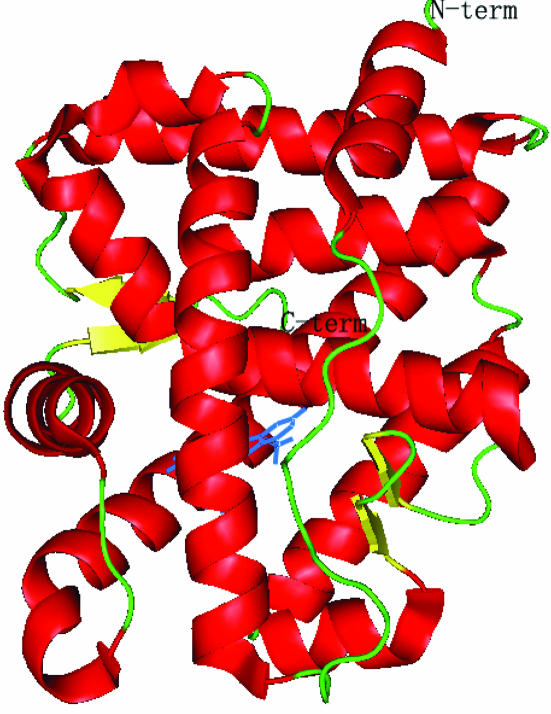
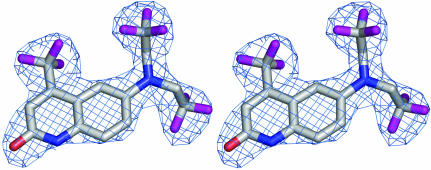
As expected, the overall structure of the human AR LBD–LGD2226 complex is very similar to that of the native model and we obtained a good electron-density map of the protein from Cys669 to Gly918 including the residues in the loop between 845 and 850 to 2.1 Å resolution. The two protein backbones are superposable. Fig. 1 ▶ shows the structure of the AR LBD–LGD2226 complex and the electron density for LGD2226 is shown in Fig. 2 ▶.
Figure 1.
A cartoon view of the AR LBD–LGD2226 complex structure, showing the position of LGD2226 (shown as a stick model).
Figure 2.
The 2F o − F c electron density of LGD2226 complexed with AR LBD calculated after final refinement. The map (contoured at the 1.5σ level) was generated using PyMOL.
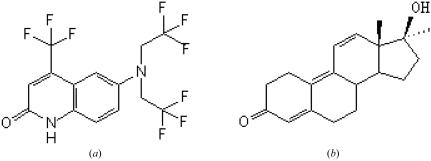
LGD2226 is a nonsteroidal SARM (van Oeveren et al., 2006 ▶) which embeds in the ligand-binding pocket in the same position as R1881 in the AR LBD–R1881 complex. The chemical structures of LGD2226 and R1881 are shown in Fig. 3 ▶. LGD2226 and R1881 share the same tropism and the two N-linked trifluoroethyl groups occupy a similar space to the C and D rings of R1881, as shown in Fig. 4 ▶.
Figure 3.
Chemical structures of LGD2226 and R1881. (a) The chemical structure of LGD2226 [6-N,N-bis(2,2,2-trifluoroethyl)amino-4-trifluoromethyl-2-quinolinone]. (b) The chemical structure of R1881 (17-methyloestra-4,9,11-trien-3-one,17b-ol).
Figure 4.
Superposition comparison between the ligands LGD2226 and R1881 in AR LBD–ligand complexes. Part of the AR LBD–LGD2226 complex is shown in red; part of the AR LBD–R1881 complex is shown in blue. Proteins are shown in trace representation; the ligands LGD2226 and R1881 are shown in stick representation.
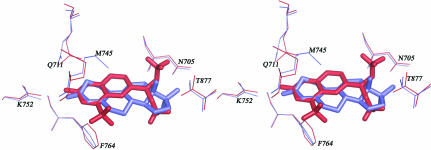
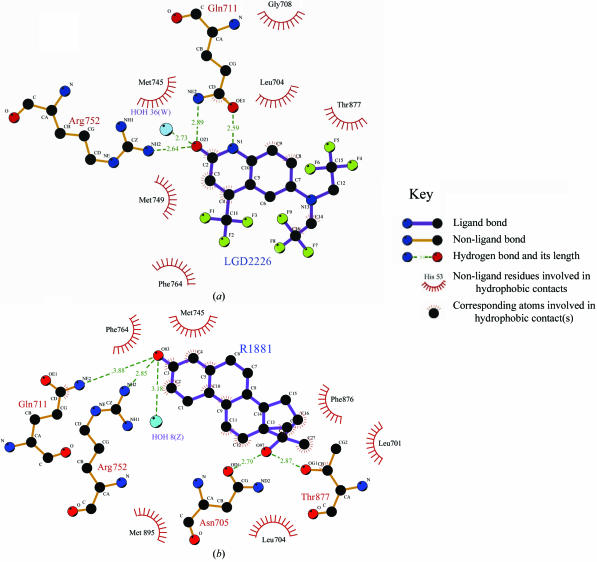
The quinolinone O atom of LGD2226 forms three hydrogen bonds with Gln711 NH2, Arg752 NH2 and a water molecule (W36); the lengths of the bonds are 2.89, 2.64 and 2.73 Å, respectively (Fig. 5 ▶ a). In the AR LBD–R1881 complex, the distances from the corresponding O atom O83 to the same three atoms are 3.88 Å (Gln711 NH2), 2.85 Å (Arg752 NH2) and 3.18 Å (water Z8) (Matias et al., 2000 ▶). The H atom on N1 of LGD2226 formed an additional hydrogen bond to Gln711 OE1. O97 of R1881 forms a hydrogen bond with Asn705 OD1 with a distance of 2.79 Å and a hydrogen bond with Thr877 OG1 with a distance of 2.87 Å (Fig. 5 ▶ b), whereas LGD2226 makes no further hydrogen bonds with AR LBD.
Figure 5.
Comparison of the interactions in the AR LBD–LGD2226 and AR LBD–R1881 complexes. (a) Detailed stereoview of the LGD2226-binding site in AR LBD. The bound LGD2226 is shown as a stick model; its surrounding residues are shown as lines and the nearby water molecule is shown as a sphere. (b) Schematic atomic interaction map of R1881 with AR LBD.
We think that the hydrogen bonds from the quinolinone O atom anchor the ligands to the receptor in both the AR LBD–LGD2226 and AR LBD–R1881 complexes. The additional hydrogen bond between N1 of LGD2226 and Gln711 OE1 helps to strengthen these other hydrogen bonds and makes them much shorter in the AR LBD–LGD2226 complex than in the AR LBD–R1881 complex. These hydrogen interactions may play a determining role in the tight packing of LGD2226 in the pocket. At the same time, the trifluoroethyl groups do not participate in hydrogen-bonding interactions. However, these substituents have a prominent influence on the biological activity in the cell-based assay. Despite the loss of two hydrogen bonds, it may be that the size or the volume of the substituent group rather than the strength of the hydrogen-bonding interactions has the more important effect on the interaction between the ligand and receptor in the AR LBD–LGD2226 complex. Since no marked changes in protein structure were observed when compared with AR LBD–R1881, it is possible that the above minute changes in interaction mode may help in further understanding the tissue-selectivity differences between SARMs.
In this communication, we have presented the crystal structure of the AR LBD–LGD2226 complex and compared it with the structure of the AR LBD–R1881 complex. LGD2226 has been proven to be a selective androgen receptor modulator with reduced effects on prostate compared with muscle and the structural information described here provides further details of its ligand-binding characteristics and provides some new insight into its ligand-binding requirements, which may aid in the discovery of more selective and effective potential therapeutic agents.
Supplementary Material
PDB reference: AR LBD–LGD2226, 2hvc, r2hvcsf
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Collins, L. L., Lee, H. J., Chen, Y. T., Chang, M., Hsu, H. Y., Yeh, S. & Chang, C. (2003). Cytogenet. Genome Res.103, 299–301. [DOI] [PubMed] [Google Scholar]
- Gao, W., Bohl, C. E. & Dalton, J. T. (2005). Chem. Rev.105, 3352–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb, B., Lombroso, R. & Beitel, L. K. (2005). Reprod. Biomed. Online, 10, 42–48. [DOI] [PubMed] [Google Scholar]
- Hamann, L. G., Higuchi, R. I., Zhi, L., Edwards, J. P., Wang, X. N., Marschke, K. B., Kong, J. W., Farmer, L. J. & Jones, T. K. (1998). J. Med. Chem.41, 623–639. [DOI] [PubMed] [Google Scholar]
- Heinlein, C. A. & Chang, C. (2002). Mol. Endocrinol.16, 2181–2187. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst.. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G. & Jones, T. A. (1998). Acta Cryst. D54, 1119–1131. [DOI] [PubMed] [Google Scholar]
- Labrie, F. (1993). Cancer, 72, 3816–3827. [DOI] [PubMed] [Google Scholar]
- Matias, P. M., Donner, P., Coelho, R., Thomaz, M., Peixoto, C., Macedo, S., Otto, N., Joschko, S., Scholz, P., Wegg, A., Basler, S., Schafer, M., Egner, U. & Carrondo, M. A. (2000). J. Biol. Chem.34, 26164–26171. [DOI] [PubMed]
- Murshudov, G. N, Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Oeveren, A. van, Motamedi, M., Mani, N. S., Marschke, K. B., Lopez, F. J., Schrader, W. T., Negro-Vilar, A. & Zhi, L. (2006). Submitted. [DOI] [PubMed]
- Pelletier, G. (2000). Histol. Histopathol.15, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Rosen, J. & Negro-Vilar, A. (2002). J. Musculoskelet. Neuronal. Interact.3, 222–224. [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995). Protein Eng.8, 127–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AR LBD–LGD2226, 2hvc, r2hvcsf