Table 1. Crystal data and data-collection statistics of crystals grown at 292 K.
Values in parentheses are for the highest resolution shell. dsDNA, double-stranded DNA. r.a., rotating-anode generator. n.d., not determined.
| Crystal form | ExoAf-I | ExoAf-II | ExoAf-III | ExoAf-IV | ExoMt-I | ExoMt-II |
|---|---|---|---|---|---|---|
| Figure | 1(a) | 1(b) | 1(c) | 1(d) | 1(e) | 1(f), 1(g) |
| Reservoir solution | 10–12% PEG 8000, 0.05 M sodium cacodylate pH 5.0, 5 mM CaCl2 | 30% PEG 4000, 0.1 M Tris pH 8.0–8.5 | 30% PEG 4000, 0.2 M Li2SO4, 0.1 M Tris pH 8.5 | 20–26% PEG 3350, 10% 2-propanol, 0.1 M Tris pH 8.0–9.5 | 10% PEG 8000, 5–8% ethylene glycol, 0.1 M NaOAc pH 5.0–5.4 | 5–10% PEG 3350, 10–15% 2-propanol, 0.1 M MES pH 5.0–6.5 |
| Protein solution | 37 mg ml−1 ExoMt, 20 mM MES pH 6.0, 0.6 M NaCl | 17 mg ml−1 (0.57 mM) ExoAf, 0.53 mM dsDNA, 9 mM MES pH 6.0, 0.28 M NaCl | 17 mg ml−1 (0.57 mM) ExoAf, 0.53 mM dsDNA, 9 mM MES pH 6.0, 0.28 M NaCl | 16.5 mg ml−1 (0.55 mM) ExoAf, 0.50 mM dsDNA, 9 mM MES pH 6.0, 0.27 M NaCl | 8 mg ml−1 (0.27 mM) ExoMt, 0.25 mM dsDNA, 15 mM MES pH 5.7, 0.38 M NaCl | 8 mg ml−1 (0.27 mM) ExoMt, 0.25 mM dsDNA, 15 mM MES pH 5.7, 0.38 M NaCl |
| Cryocooling condition | — (298 K) | — (298 K) | Straight from drop | Paraffin oil | 25%(v/v) Ethylene glycol | — (298 K) |
| X-ray source | r.a. | r.a. | r.a./BESSY BL14.2 | r.a. | r.a./BESSY BL14.1 | r.a. |
| Wavelength (Å) | 1.5418 | 1.5418 | 1.5418/0.9537 | 1.5418 | 1.5418/0.9537 | 1.5418 |
| Data-collection temperature (K) | 298 | 298 | 298/100 | 100 | 298/100 | 298 |
| Space group† | P6x | P2x | P21 | P6x | P21 | P4x |
| Resolution limit (Å) | 2.5 | 3.2 | 2.2/1.7‡ | 2.3 | 3.0/1.8‡ | 3.0 |
| Unit-cell parameters | ||||||
| a (Å) | 252.7 | 84.5 | 69.6/67.8 | 92.0 | 45.0/44.4 | 101.3 |
| b (Å) | 252.7 | 77.9 | 61.3/60.8 | 92.0 | 99.3/96.8 | 101.3 |
| c (Å) | 76.2 | 89.0 | 77.5/72.4 | 89.0 | 81.5/80.8 | 80.7 |
| β (°) | 92.3 | 95.0/91.2 | 97.8/97.8 | |||
| VM§ (Å3 Da−1) | 2.0–3.9 | 2.0–3.3 | 2.5/2.3¶ | 3.6 | 2.7/2.6¶ | 2.3 or 3.4 |
| Solvent content§ (%) | 37–68 | 37, 50 or 62 | 51/46¶ | 66 | 54/52¶ | 46 or 64 |
| Molecules per ASU§ | 12–6 | 5–3 | 2¶ | 1 | 2¶ | 3–2 |
| Total reflections | n.d. | n.d. | 507681 | n.d. | 306722 | n.d. |
| Unique reflections | n.d. | n.d. | 64871 | n.d. | 61516 | n.d. |
| Completeness†† | n.d. | n.d. | 100 (99.9) | n.d. | 98.2 (96.6) | n.d. |
| Rsym††‡‡ | n.d. | n.d. | 7.0 (43.6) | n.d. | 6.0 (48.2) | n.d. |
| Rp.i.m.††§§ | n.d. | n.d. | 2.2 (20.5) | n.d. | 2.9 (24.3) | n.d. |
x, possible screw axes could not be determined owing to insufficient data.
Unit-cell parameters, Matthews coefficient and related values and data-collection statistics correspond to the data sets at the two given temperatures.
Matthews coefficient (Matthews, 1968 ▶) and related values were calculated under the presumption that no DNA had bound to the protein.
Values derived from the presence of one DNA molecule per ASU in addition to protein.
Data-processing statistics are only given for synchrotron data collected at 100 K.
R
sym = 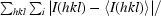
 .
.
R
p.i.m. = 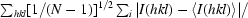
 . R
p.i.m. describes the precision of merged reflections by incorporation of the multiplicity N (Weiss, 2001 ▶).
. R
p.i.m. describes the precision of merged reflections by incorporation of the multiplicity N (Weiss, 2001 ▶).
