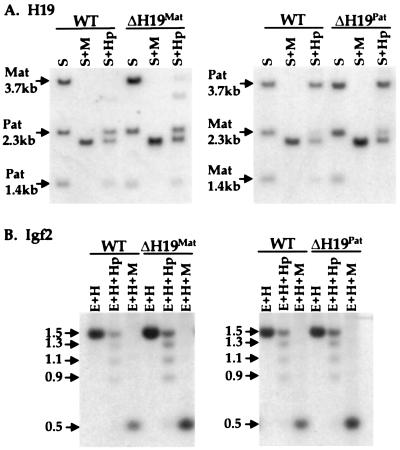
Figure 5.
Methylation of H19 and Igf2 in loxΔH19 heterozygotes. (A) Methylation analysis of the H19 5′ flanking region. DNA from wild-type (WT) mice and mice carrying loxΔH19Mat (Left) or loxΔH19Pat (Right) alleles was digested with SacI (S) and SacI plus HpaII (Hp) or MspI (M). The blots were hybridized to a 3.8-kb EcoRI probe from the 5′ flank of the H19 gene. The partially methylated maternal H19 DNA in loxΔH19Mat animals is more apparent than in wild-type animals because of loading differences. (B) Methylation analysis of the Igf2 DMR1 region. Liver genomic DNAs from the same animals as in A were digested with EcoRI (E) and HindIII (H) or with EcoRI–HindIII plus HpaII or MspI. The blots were hybridized to a 717-bp probe from the DMR1 region of the Igf2 5′ flank. In these animals, the 1.5-kb band represents the fully methylated paternal allele whereas the reduced methylation on the maternal allele gives a range of smaller digestion products.