Figure 7. .
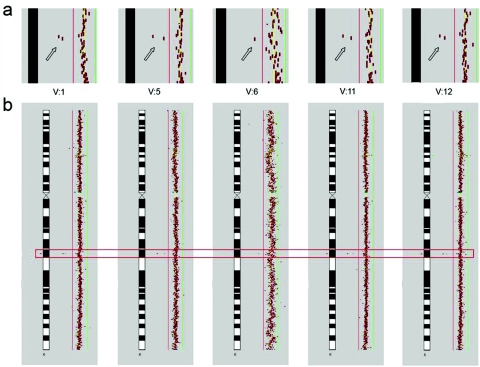
Array CGH results for patients with MR. Submegabase-resolution array CGH was performed as described elsewhere.15 After normalization by subgrid LOWESS, Cy3:Cy5 intensity ratios of each clone were plotted in a size-dependent manner along the chromosome ideograms, by use of CGHPRO.16 Red and green bars indicate the log2 ratio thresholds of −0.3 (loss) and 0.3 (gain), respectively. A homozygous deletion of 6q16.3 was the only aberration that cosegregated with the disease; it was present in all affected probands and was absent in all unaffected probands tested. Array CGH data shown here have been deposited in NCBI Gene Expression Omnibus (GEO)17 and are accessible through GEO series accession number GSE 7886. a, Close-up of findings for patient 6q16.3, demonstrating a deletion of two BAC clones (RP11-226C08 and RP11-543M18 [arrows]) encompassing GRIK2. (Because of lower hybridization quality for V:6, only one BAC represents the DNA copy-number loss.) b, Overall view of chromosome 6 in the corresponding patients. A red rectangle highlights the region depicted in panel a.