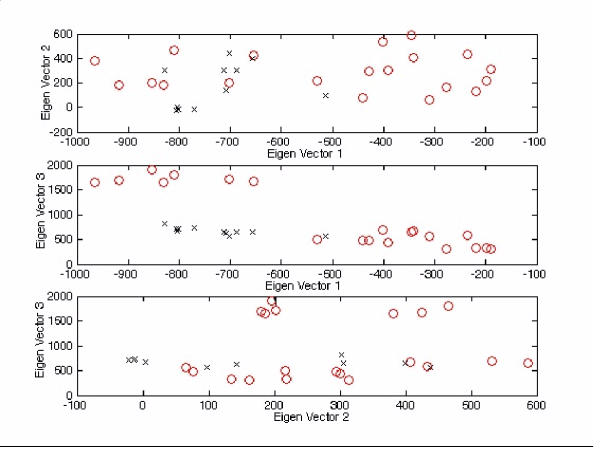
Figure 6.
2-D plots for Figure 5 with different viewpoint specifications. The tetra-nucleotide usage patterns table in the additional file 2 (entire genome) for each virus have been redisplayed on the (1st vs 2nd), (1st vs 3rd) and (2nd vs 3rd) eigen-vector axes ('o' represents positive strand ssRNA virus, 'x' represents negative strand ssRNA virus). For the top figure, the order for 'o' is [3,7,1,4,2,5,6,17,13,20,10,16,9,8,11,15,12,14,18,19]* (left to right), whereas 'x' is [26,22,30,23,24,28,31,27,25,29,21]* (left to right). For the middle figure, the order for 'o' is [3,7,1,4,2,5,6,17,13,20,10,16,9,8,11,15,12,14,18,19]* (left to right), whereas 'x' is [26,22,30,23,24,28,31,27,25,29,21]* (left to right). For the bottom figure, the order for 'o' is [3,7,1,4,2,5,6,17,13,20,10,16,9,8,11,15,12,14,18,19]* (left to right), whereas 'x' is [26,22,30,23,24,28,31,27,25,29,21]* (left to right). *The corresponded virus for each number follows Figure 5.