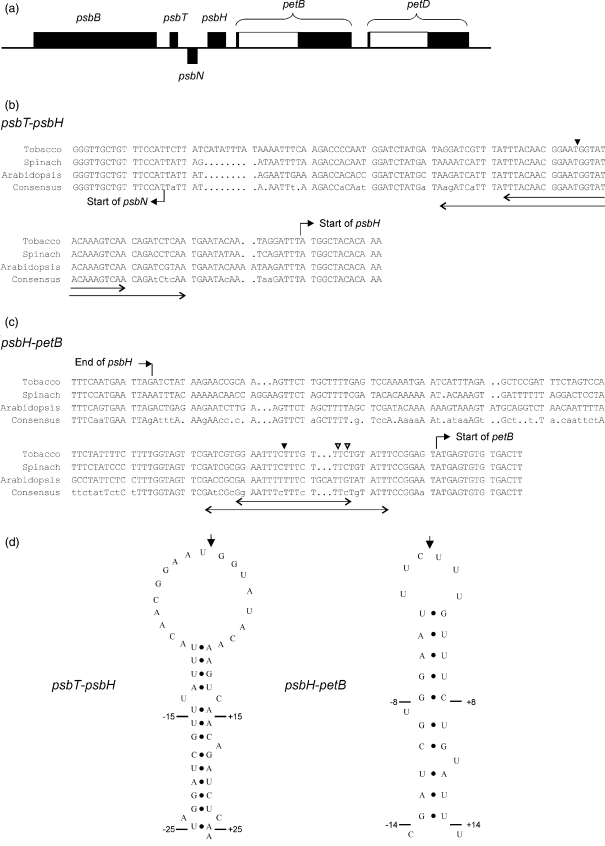
Figure 1. Identification of intercistronic mRNA processing sites in the tobacco psbB operon.
(a) Structure of the psbB operon. Genes above the lines are transcribed from left to right; the gene below the line (psbN) is transcribed in the opposite direction. The group II introns within the petB and petD coding regions are shown as open boxes. Transcription from the psbB promoter produces a pentacistronic mRNA that undergoes a complex series of processing steps resulting in monocistronic and oligocistronic mRNA species (Westhoff and Herrmann, 1988).
(b) Partial sequence alignment of the psbT–psbH spacer region from tobacco, spinach and Arabidopsis. Shown is the 3′ part of the spacer, between the antisense psbN sequence and psbH. The intercistronic RNA processing site mapped in tobacco is indicated by a closed triangle. The sequences chosen as putative processing sequences in plastid transformation experiments are indicated by the double-arrowed lines.
(c) Alignment of the psbH–petB spacer regions from tobacco, spinach and Arabidopsis. The major intercistronic RNA processing site mapped in tobacco is marked by a closed triangle; additionally identified minor processing sites are indicated by open triangles.
(d) Location of intercistronic processing sites within putative RNA stem–loop structures. The major endonucleolytic cleavage sites are indicated by arrowheads.