Figure 2. Generation of plastid-transformed plants to test putative intercistronic processing elements in vivo.
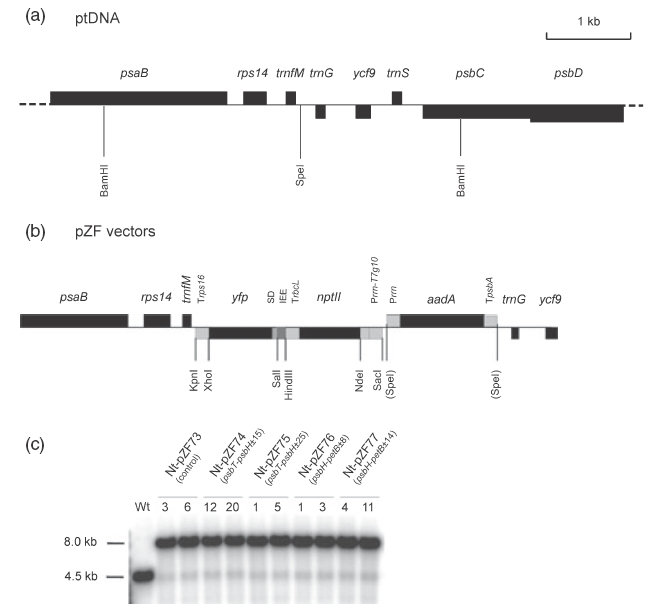
(a) Map of the targeting region in the tobacco plastid genome (ptDNA). Genes above the lines are transcribed from left to right; genes below the line are transcribed in the opposite direction. The transgenes are targeted to the intergenic spacer between the trnfM and trnG genes.
(b) Construction of plastid transformation vectors (pZF series) integrating an operon of two transgenes (nptII and yfp) into the tobacco plastid genome, along with the selectable marker gene for chloroplast transformation, aadA (Svab and Maliga, 1993). Relevant restriction sites used for cloning and RFLP analysis are indicated, sites lost by ligation of heterologous ends are shown in parentheses. Prrn, rRNA operon promoter; TpsbA, 3′ UTR of the psbA gene; Prrn-T7g10, rRNA operon promoter fused to the leader sequence of bacteriophage T7 gene 10; TrbcL, 3′ UTR of the rbcL gene; IEE (putative processing element); SD, Shine–Dalgarno sequence; Trps16, 3′ UTR of the rps16 gene.
(c) Southern blot confirming plastid transformation and assessing homoplasmy of transplastomic lines. Digestion with BamHI produces fragments of approximately 4.5 kb in the wild-type and approximately 8 kb in all transplastomic lines. This size difference corresponds exactly to the combined size of the three integrated transgenes. Note that the probe (a radiolabeled ycf9 fragment) detects a faint wild-type-like band in all transplastomic lines that has been shown previously to come from promiscuous chloroplast DNA in the tobacco nuclear (or mitochondrial) genome (Ruf et al., 2000).
