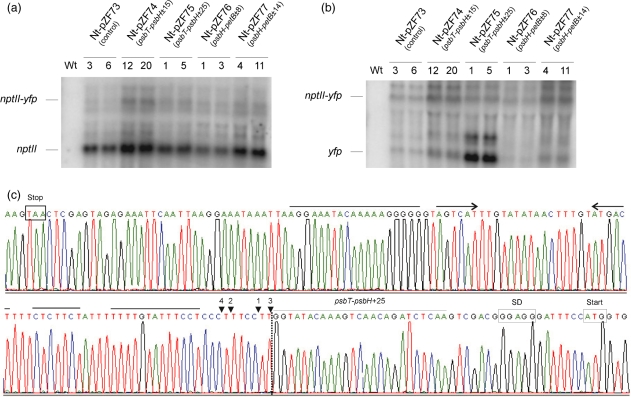
Figure 4. RNA accumulation in transplastomic lines harboring various candidate processing elements between the nptII and yfp cistrons.
(a) Accumulation of nptII mRNA. All transplastomic lines accumulate predominantly monocistronic nptII mRNA, in addition to small amounts of di-cistronic nptII–yfp transcripts and other minor RNA species that were not further characterized. Accumulation of monocistronic nptII message in the Nt-pZF73 control lines demonstrates that 3′ processing of nptII mRNA is independent of the presence of the putative processing sequences.
(b) Accumulation of yfp mRNA. Significant amounts of (monocistronic) yfp mRNA accumulated only in the Nt-pZF75 lines that harbor the ±25 IEE from the psbT–psbH intergenic spacer. Note that, in addition to the monocistronic yfp and di-cistronic nptII–yfp transcripts, two minor RNA species also accumulate. One of them is approximately 200 bp larger than the monocistronic yfp message, and the other is approximately 200 bp larger than the di-cistronic nptII–yfp transcript. These minor RNA species were not further characterized, but the most probable explanation is that they originate from read-through transcription through trnfM (see Figure 1b), whose antisense transcript can also fold into a stable cloverleaf-like secondary structure and thus act as an RNA processing signal.
(c) Mapping of the 5′ and 3′ ends of the monocistronic yfp mRNA in Nt-pZF75 plants. The sequence of a cDNA clone derived from head-to-tail ligated yfp mRNA is shown. The ligation site (i.e. the border between the 3′ end and the 5′ end of the circularized mRNA) is indicated by the dotted vertical line. Note that the 5′ end generated by processing within the psbT–psbH spacer element is identical in all ten cDNA clones and corresponds precisely to the 5′ end of the psbH mRNA (Figure 1b,d). Alternative 3′ ends are indicated by arrowheads, and the number of clones in which the respective termini were found is indicated. A putative transcript-stabilizing stem–loop-type RNA secondary structure within the rps16 3′ UTR is marked by horizontal arrows (interruptions indicate unpaired nucleotides). The stop codon, Shine–Dalgarno sequence and start codon are boxed.