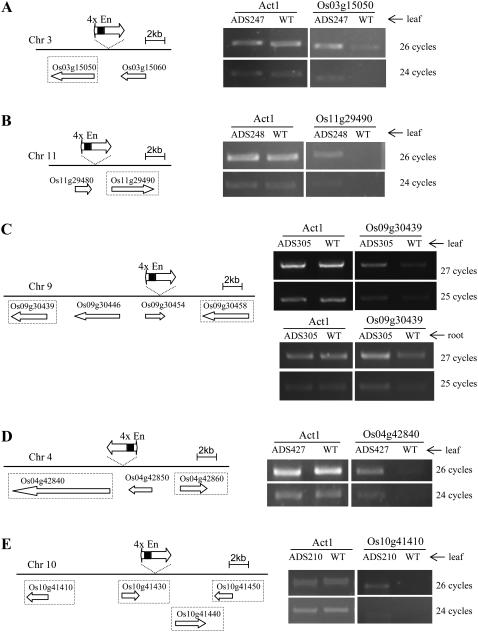
Figure 4.
Semiquantitative RT-PCR analysis of five Ds insertion lines. Left, Schematic representation of the Ds insertion and the CaMV 35S enhancers (4xEn) relative to the nearby genes. The map of each chromosomal region was based on the Rice GE genome database RiceGE Functional Genomics Database (http://signal.salk.edu/cgi-bin/RiceGE). Genes that were examined in semiquantitative RT-PCR are marked with dotted-line boxes. Right, RT-PCR analysis of activation-tagged genes. Rice Act1 transcript was amplified as control. WT, Wild-type rice ‘Nipponbare’. RT-PCR using leaf or root RNA was indicated. A, Line ADS247 was tagged by the activation-tagging Ds element 1,609 bp upstream of Os03g15050, which encodes the phosphenolpyruvate carboxykinase. B, Line ADS248 was tagged by a Ds 1,880 bp upstream of Os11g29490, which encodes a plasma membrane-type ATPase. C, Line ADS305 was tagged by a Ds 12.2 kb upstream of Os09g30439, which encodes a heat shock protein. D, Line ADS427 was tagged by a Ds 1,349 bp upstream of Os04g42840, which encodes a HEAT repeat family protein. E, Line ADS210 was tagged by a Ds 11.8 kb upstream of Os10g41410, which encodes the nucleoside diphosphate kinase 1. For each gene, the RT-PCR result was confirmed using two different pairs of specific primers and the result from using one pair of primers was shown. Each PCR experiment was repeated at least three times to confirm the result.