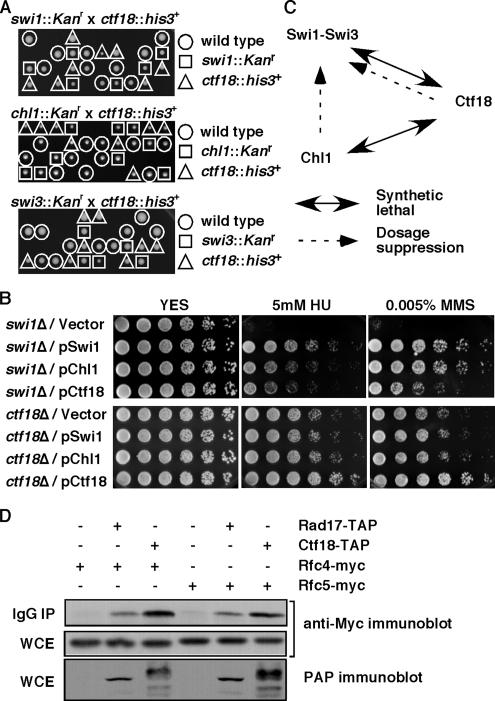
Figure 1.
Genetic interaction involving Swi1-Swi3, RFCCtf18, and Chl1. (A) None of the viable spores from swi1::Kanr x ctf18::his3+, chl1::Kanr × ctf18::his3+, and swi3::Kanr × ctf18::his3+ crosses were able to grow on YES medium containing G-418 and EMM2, Edinburgh minimal media lacking histidine, indicating that swi1Δ ctf18Δ, chl1Δ ctf18Δ, and swi3Δ ctf18Δ cells are inviable. Genotypes of viable spores are shown. Representative images of >40 tetrad dissections from each cross are shown. (B) Damage sensitivities of swi1Δ were suppressed by an increased gene dosage of chl1+ or ctf18+. swi1Δ or ctf18Δ cells were transformed with the indicated plasmid and plated on YES medium containing no drug (YES), 5 mM HU, or 0.005% MMS. Fivefold serial dilution of cells were plated and incubated for 2–3 d at 32°C. Representative images of repeat experiments are shown. (C) Summary of genetic interaction involving Swi1-Swi3, RFCCtf18, and Chl1. (D) Ctf18 associates with Rfc4 and Rfc5. Protein extracts were prepared from cells expressing the indicated fusion proteins. Ctf18-TAP was precipitated and probed with anti-myc antibodies. Rfc4-Myc and Rfc5-Myc are shown to be equally expressed in each cell line (whole-cell extract, WCE). As a positive control, Rad17-TAP is shown to associate with Rfc4 and Rfc5. IgG IP, precipitated fraction.