Abstract
Polyploidy is known to induce numerous genetic and epigenetic changes but little is known about their physiological bases. In wheat, grain texture is mainly determined by the Hardness (Ha) locus consisting of genes Puroindoline a (Pina) and b (Pinb). These genes are conserved in diploid progenitors but were deleted from the A and B genomes of tetraploid Triticum turgidum (AB). We now report the recurrent deletions of Pina-Pinb in other lineages of polyploid wheat. We analyzed the Ha haplotype structure in 90 diploid and 300 polyploid accessions of Triticum and Aegilops spp. Pin genes were conserved in all diploid species and deletion haplotypes were detected in all polyploid Triticum and most of the polyploid Aegilops spp. Two Pina-Pinb deletion haplotypes were found in hexaploid wheat (Triticum aestivum; ABD). Pina and Pinb were eliminated from the G genome, but maintained in the A genome of tetraploid Triticum timopheevii (AG). Subsequently, Pina and Pinb were deleted from the A genome but retained in the Am genome of hexaploid Triticum zhukovskyi (AmAG). Comparison of deletion breakpoints demonstrated that the Pina-Pinb deletion occurred independently and recurrently in the four polyploid wheat species. The implications of Pina-Pinb deletions for polyploid-driven evolution of gene and genome and its possible physiological significance are discussed.
For over 100 years, wheat grain has been classified into hard and soft types. Grain hardness or texture is mainly determined by the Hardness (Ha) locus. This classification forms the fundamental basis for differentiating wheat grain worldwide (for review, see Morris, 2002). Wheat speciation has been molded by polyploidy. Diploid (Triticum monococcum), tetraploid (Triticum turgidum), and hexaploid (Triticum aestivum) wheat species have been known since the 1920s (for review, see Gill and Friebe, 2002). Of the 600 million metric tons of wheat produced in the world in 2005 (http://faostat.fao.org), over 90% comes from hexaploid wheat, also called common or bread wheat, and the remaining from tetraploid, also called macaroni or durum wheat (http://www.fas.usda.gov/pecad/highlights/2005/10/durum_27oct2005). Bread wheat grain is either soft and used for pastries or hard and used for bread and noodles. Durum wheat grain is classified as extrahard and used for pasta. Diploid wheat grain is soft and, although it was the first domesticated wheat, is now grown as a specialty crop in isolated areas. Recently, See et al. (2004) produced a supersoft hexaploid wheat genotype by introgressing softness genes from diploid species.
Because of the pivotal importance of grain texture in determining end use quality, this trait has been intensively studied by geneticists, cereal chemists, and, more recently, by molecular biologists. In the 1970s, Mattern et al. (1973) mapped a gene with a major effect on grain texture on chromosome 5D. Later, Law et al. (1978) further localized the gene to the short arm of chromosome 5D and designated the trait as hardness with alleles Ha for soft and ha for hard. Greenwell and Schofield (1986) found that a 15-kD protein called friabilin from water-washed starch from grain was associated with the Ha locus. Abundant friabilin was found in soft wheat with Ha alleles, small amounts in hard wheat with ha allele, and none in extrahard durum wheat grains. Blochet et al. (1991, 1993) isolated and sequenced lipid-binding proteins and called them puroindolines because of the presence of a Trp domain. Gautier et al. (1994) isolated cDNA clones corresponding to genes Puroindoline a (Pina) and b (Pinb). A large body of research has shown that PINA, PINB, and grain softness protein (GSP) constitute a major fraction of friabilin. Functional copies of both Pin genes are required for soft grain texture in wheat (for review, see Morris, 2002).
Cloning of the Pin genes stimulated genomics research on the Ha locus. Gautier et al. (2000), using a PCR approach, showed that Pina and Pinb are highly conserved in diploid wheat and the Triticeae and closely related cereals, such as rye (Secale cereale), barley (Hordeum vulgare), and oats (Avena sativa), but were absent in sorghum (Sorghum bicolor), maize (Zea mays), and rice (Oryza sativa). They were reported absent in tetraploid wheat species T. turgidum and Triticum timopheevii and reintroduced into hexaploid wheat as it arose from the hybridization of tetraploid T. turgidum (lacking Pin genes) and Aegilops tauschii Coss. (Pin genes present). Null mutations of PINA protein in hexaploid wheat cultivars of more recent origin have been reported (for review, see Morris, 2002).
Sequencing the Ha loci of T. monococcum (Chantret et al., 2004) and A. tauschii (Chantret et al., 2005) showed that Gsp (Gene2), Pina (Gene4), and Pinb (Gene6) are located in an interval of approximately 70 kb. A hypothetical gene (Gene3) was found between Gsp and Pina and a function-unknown gene (Gene5) between Pina and Pinb. In the D genome of A. tauschii and T. aestivum, a 5′ untranslated region and a 5′ coding sequence of Pinb were duplicated downstream of the functional Pinb. Upstream of Gsp, BGGP (Gene1), coding for a β-1-3-galactosyl-O-glycosyl-glycoprotein, delimits the 5′ boundary of Ha. Downstream of Pinb, a cluster of ATPase genes (Gene7) coding for AAA-type ATPase and an unknown gene (Gene8) delimit the 3′ boundary of Ha. Compared to Ha-D, deletions of the genomic block containing Gene3, Pina, Gene5, and Pinb occurred at Ha-A and Ha-B of T. turgidum and T. aestivum (Chantret et al., 2005).
During routine mapping of the tetraploid T. timopheevii genome, we detected one copy of the Pin genes in its genome. This was a surprising result in view of the previous report of Gautier et al. (2000) and could be due to polymorphism for this locus in this species. A large and more comprehensive polymorphism survey of Pin genes in polyploid wheat species was initiated. To more fully characterize the nature of deletions for this region in polyploid wheat, we undertook sequencing of the Ha region of Aegilops speltoides Tausch., of which the S genome has highest affinity among the diploid species to the B and G genomes of polyploid wheat. Because two of the three genomes of polyploid wheat were donated by Aegilops spp., we also analyzed haplotype structure at the Ha locus of a small number of diploid and polyploid Aegilops spp. These results and their implications for polyploidy-driven mechanisms of gene and genome evolution and speciation are reported and their possible physiological significance is discussed.
RESULTS
Phylogeny of Polyploid Wheat and Genetic Nomenclature of Loci
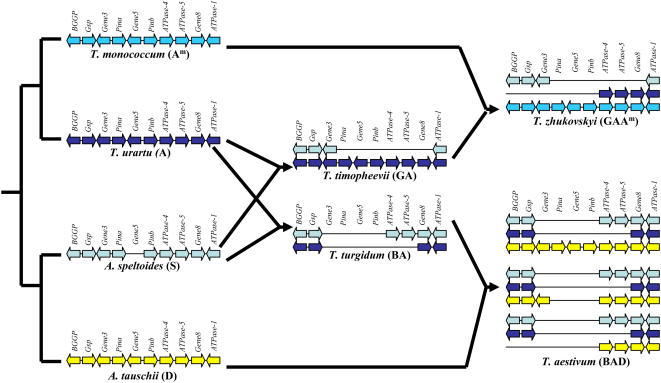
To interpret the Ha deletion haplotype survey results, it is important to briefly introduce understanding of the phylogeny of polyploid wheat. Two lineages of tetraploid wheat, emmer (T. turgidum, 2n = 4x = 28; AB) and Timopheevi (T. timopheevii Zhuk., 2n = 4x = 28; AG), originated less than 0.5 million years ago (Huang et al., 2002) from two separate hybridization events between A. speltoides (2n = 2x = 14; S), as the female parent, and Triticum urartu Tumanian ex Gandilyan (2n = 2x = 14; A), as the male parent (Tsunewaki, 1993; Dvorak, 1998; Kilian et al., 2007). More recent hybridizations with two additional diploid species gave rise to hexaploid wheat lineages. Common wheat (T. aestivum; ABD) originated approximately 8,000 years ago (Nesbitt and Samuel, 1996) from hybridization between T. turgidum and A. tauschii in cultivated fields and does not exist in the wild (Kihara, 1944; McFadden and Sears, 1946). Triticum zhukovskyi Menabde and Ericzjan (2n = 6x = 42; AAmG) arose in cultivation from hybridization of T. timopheevii sp. timopheevii with T. monococcum L. sp. monococcum (2n = 2x = 14; Am; Upadhya and Swaminathan, 1963; Dvorak et al., 1993). The A, B, and D diploid donors of polyploid wheat diverged from a common ancestor approximately 3 million years ago (Huang et al., 2002). In polyploid wheat, the Ha-A, Ha-B, and Ha-D genetic nomenclature conveys both locus and genomic origin of Ha homoeoloci. Results summarizing the haplotype structure at the Ha locus in relation to the phylogeny of polyploid wheat are shown in Figure 1.
Figure 1.
Scheme for polyploid wheat phylogeny and changes in the Ha haplotype structure during their evolution. Solid arrows represent genes and their orientation, with symbols above. Species and their genome formulae are indicated underneath. Arrow size and spacing are not proportional to gene size or intergenic interval.
Sequence Analysis of the Ha-S Genomic Region of A. speltoides
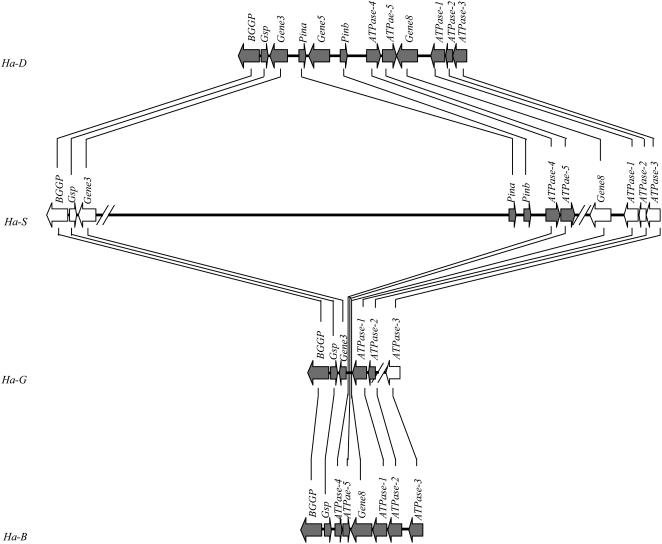
Of the three diploid ancestors of polyploid wheat, bacterial artificial chromosome (BAC) sequences of the Ha genomic region of A- and D-genome ancestors were reported previously (Chantret et al., 2004, 2005). We undertook sequence analysis of a BAC from the S genome of A. speltoides. We screened a BAC library of A. speltoides (Akhunov et al., 2005) with Gsp and Pina probes, each of which identified three BACs. Gsp-containing BACs did not overlap with Pina-containing BACs. We estimate that the Ha-S genomic region in A. speltoides is 3 times the size of Ha-Am in T. monococcum and of Ha-D in A. tauschii and 5 times the size of Ha-D of T. aestivum. A Pina BAC, 197O23, was shotgun sequenced at 8× coverage and assembled into 13 contigs after prefinishing, totaling 212,510 bp. Four nontransposable element (TE) protein-coding genes, Pina, Pinb, and two ATPases, were found in this BAC, are located in the contig at the 3′ end in the same orientation, and span 28,848 bp (Fig. 2). Gene5, previously reported to be present in the collinear region between Pina and Pinb of T. monococcum and A. tauschii, was not found in A. speltoides. Based on sequence homology and collinearity, the two ATPase genes in BAC 197O23 correspond to ATPase-4 and ATPase-5 at the Ha-D locus and are orthologous to two truncated ATPase genes upstream of Gene8 at the Ha-B locus (Chantret et al., 2005). The rest of BAC 197O23 is gene free and mainly occupied by TEs and tandem repeats, a typical feature of large genomes, where genes are clustered into islands and separated by nested TEs (Wicker et al., 2001). Gene8, ATPase-1, ATPase-2, and ATPase-3 were located in a separate BAC.
Figure 2.
Comparison of haplotype structure at the Ha loci: Ha-D of A. tauschii and T. aestivum, Ha-S of A. speltoides, Ha-G of T. timopheevii, and Ha-B of T. turgidum and T. aestivum. Solid arrows represent genes and their orientation with the gene symbols above them. Gene3 and Gene8 are hypothetical and Gene5 is coding for an unknown protein. The Ha-S haplotype spans three separate BACs as indicated by slashes. White arrows in Ha-S and Ha-G were not sequenced; they are deduced based on collinearity between the Ha-B and Ha-D loci. The lines connect the orthologs. The Ha-B and Ha-D haplotypes are after Chantret et al. (2005).
Survey of the Haplotype Structure at the Ha Locus in Aegilops and Triticum
We determined the haplotype structure at the Ha locus by Southern analysis of tester DNA digested with restriction enzymes EcoRI, HindIII, or BamHI using Pina and Pinb gene probes. We estimated the copy number of Pina or Pinb genes in tester species by counting the number of fragments detected by Southern hybridization. The data were tabulated to determine whether the haplotype structure was conserved or there were null haplotypes for either one or both the Pin genes at the Ha locus (Table I; Supplemental Table S1). Null haplotypes were further characterized according to the size of the deletion, either by Southern analysis using additional gene probes that mark the Ha locus (see Fig. 1), or by sequencing as described below.
Table I.
Summary of plant materials used, their ploidy levels, and their Pina-Pinb haplotypes
For Aegilops polyploids, genome locations of Pina-Pinb haplotypes were not determined.
| Species | Genome | No. of Accessions | Pina-Pinb Haplotypea | ||
|---|---|---|---|---|---|
| Diploid Triticum | A | B/G | Am/D | ||
| T. urartu | A | 20 | + + | ||
| T. monococcum | Am | 3 | + + | ||
| Tetraploid Triticum | |||||
| T. turgidum | AB | 93 | − − | − − | |
| T. timopheevii | AG | 71 | + + | − − | |
| Hexaploid Triticum | |||||
| T. aestivum | ABD | 37 | − − | − − | + + |
| 3 | − − | − − | − − | ||
| T. zhukovskyi | AGAm | 3 | − − | − − | + + |
| Diploid Aegilops | Genome 1 | Genome 2 | Genome 3 | ||
| A. tauschii | D | 4 | + + | ||
| A. speltoides | S (≈B or G) | 45 | + + | ||
| A. bicornis | Sb | 2 | + + | ||
| A. longissima | Sl | 2 | + + | ||
| A. searsii | Ss | 2 | + + | ||
| A. sharonensis | Ssh | 3 | + + | ||
| A. caudata | C | 2 | + + | ||
| A. comosa | M | 3 | + + | ||
| A. umbellulata | U | 2 | + + | ||
| A. uniaristata | N | 2 | + + | ||
| Tetraploid Aegilops | |||||
| A. crassa | DcrX | 11 | + + | + + | |
| 1 | + + | − + | |||
| A. cylindrica | DcCc | 3 | + + | + + | |
| A. ventricosa | DvN | 15 | + + | + + | |
| A. biuncialis | UM | 2 | + + | + + | |
| A. neglecta | UM | 11 | + + | + + | |
| 16 | + + | − + | |||
| A. columnaris | UcoMco | 1 | + + | + + | |
| A. geniculata | UgMg | 2 | + + | + + | |
| 1 | + + | − + | |||
| A. kotschyii | USs | 9 | + + | + + | |
| A. peregrina | USs | 1 | + + | + + | |
| A. triuncialis | UtCt | 1 | + + | + + | |
| 1 | + + | − + | |||
| Hexaploid Aegilops | |||||
| A. crassa | DDcrX | 3 | + + | + + | − + |
| 2b | + + | + + | − − | ||
| A. juvenalis | DcrXU | 4 | + + | + + | + − |
| A. vavilovii | DcrXSs | 2 | + + | + + | − + |
| 1b | + + | + + | − − | ||
| A. neglecta | UnMnNn | 6 | + + | + + | − + |
| 4 | + + | − + | − + | ||
| 3b | + + | − + | − − | ||
+ indicates presence and − indicates deletion; + +, both Pina and Pinb are present; − −, both Pina and Pinb are deleted; − +, Pina is deleted but Pinb is present; + −, vice versa.
Genomes containing the Pina deletion were not determined relative to the Pinb deletion. − − does not necessarily indicate a haplotype where both Pina and Pinb were deleted from the same genome.
Diploid Species
We randomly selected at least two accessions from each of the 12 diploid species of Aegilops and Triticum (a total of 90 accessions; Table I; Supplemental Table S1) for the haplotype survey. In all cases, Southern hybridization detected a single band or, rarely, multiple bands for Pina and Pinb gene probes, indicating that haplotype structure at the Ha locus is conserved in the diploid species. A single copy of Pina and Pinb was detected in A- and D-genome donor species of polyploid wheat (Table I). Five Aegilops species share the S genome and all except A. speltoides are self-pollinated. All self-pollinated S-genome species had one copy of Pina and Pinb. Most accessions of A. speltoides also carry one copy of the Pin genes and the observed multiple Southern hybridization fragments in some accessions (Supplemental Table S1) may be due to either heterozygosity, because it is a cross-pollinated species, or, rarely, the presence of intragenic restriction sites or gene duplication. All the C-, M-, U-, and N-genome species also had one copy of the Pin genes, except for one accession of Aegilops comosa, where Southern analysis indicated multiple gene copies.
Tetraploid Species
The above-mentioned diploid species have contributed genomes to tetraploid Triticum and Aegilops spp. and two copies each of Pina and Pinb genes are expected in the genomes of these tetraploid species (Table I). The tetraploid wheat species T. turgidum (AB) and T. timopheevii (AG) form the A-genome cluster. We screened 92 accessions of T. turgidum, including eight subspecies representing the range of wild and domesticated forms. All showed the null haplotype for the Pin genes (Table I; Supplemental Table S1; Fig. 3), confirming that Pina and Pinb have been deleted in this species (Gautier et al., 2000; Dvorak et al., 2004).
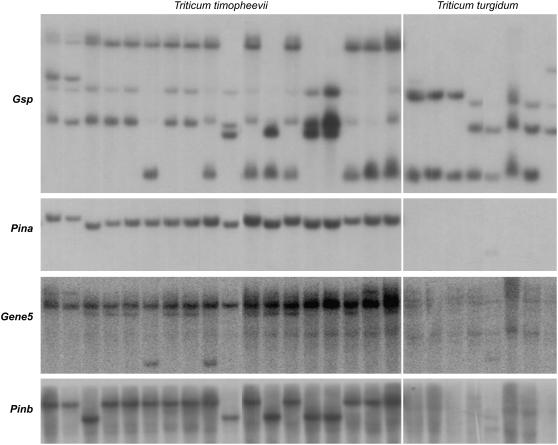
Figure 3.
The autoradiogram of a Southern hybridization. Genes Gsp, Pina, Gene5, and Pinb are indicated at the left and species at the top. Gsp detected two major bands in both T. timopheevii and T. turgidum. Pina, Gene5, and Pinb detected a single fragment in T. timopheevii and none in T. turgidum.
We screened 65 accessions of T. timopheevii, including two subspecies representing the range of wild and domesticated forms (Supplemental Table S1). All carried only one copy of the Pina and Pinb genes, indicating null haplotype at the Ha locus for one of its genomes (Supplemental Table S1). Gene5, which lies between Pina and Pinb, was, as expected, present in one copy. The Gsp probe detected two copies, indicating that one of the breakpoints that produced the null haplotype is located between Gsp and Pina (Fig. 3).
Nine tetraploid Aegilops spp. are grouped into the U- and D-genome clusters. In the U-genome cluster, one accession each was analyzed for Aegilops columnaris Zhuk. (UcoMco) and Aegilops peregrina (Hack. in J. Fraser) Marie and Weiller (USs), two accessions for Aegilops biuncialis Vis. (UM), and nine accessions for Aegilops kotschyii Boiss. (USs); all showed conserved haplotype structure for the Pin genes in both of their genomes (Table I). The other three U-genome cluster species, Aegilops geniculata Roth (UgMg), Aegilops neglecta Req. ex Bertol. (UM), and Aegilops triuncialis L. (UtCt), where more than one accession was analyzed, were polymorphic in Pina copy number. One or more accessions in each species had a conserved haplotype structure for the Pin genes and the other accessions for the same species showed unique haplotypes in one of their genomes where the Pina gene was deleted, but the Pinb gene was retained. Therefore, it is possible that, if a larger survey of the U-genome cluster species is undertaken, all may turn out to be polymorphic for Ha haplotype structure.
The D-genome cluster species Aegilops crassa Boiss. (DX), Aegilops cylindrica Host (DcCc), and Aegilops ventricosa Tausch (DvNv) showed a conserved haplotype structure for the Pin genes, except one of the 12 accessions of A. crassa was null for Pina in one of its genomes.
Hexaploid Species
Some of the tetraploid species of Triticum and Aegilops mentioned above have undergone a second round of hybridization with various diploid species to form hexaploid species. The A-genome cluster species T. aestivum (ABD) is expected to have one copy each of the Pina and Pinb genes contributed by A. tauschii (D). We surveyed 40 accessions of T. aestivum, including all six subspecies and 18 historical hard wheat cultivars of T. aestivum sp. aestivum. Of the 40 T. aestivum accessions, 37 showed a conserved haplotype structure for Pina and Pinb in the D genome (Ha-D) and, as expected, a null haplotype for the A and B genomes. Of the 18 hard wheat cultivars that do not express the PINA protein (Morris et al., 2001), ‘Red Egyptian’, ‘Sea Island’, and ‘Komar’ showed the null haplotype at Ha-D. Further probing with BGGP and Gsp detected three copies in ‘Sea Island’ and ‘Komar’, but only two in ‘Red Egyptian’ (Fig. 4). RFLP analysis of the ‘Chinese Spring’/‘Red Egyptian’ chromosome substitution lines 5A, 5B, and 5D confirmed that BGGP, Gsp, Pina, and Pinb were deleted from the Ha-D locus on chromosome 5D of ‘Red Egyptian’. Therefore, at least two independent deletion events occurred at the Ha-D locus in common wheat, one with the distal breakpoints between Gsp and Pina in ‘Sea Island’ and ‘Komar’, and another haplotype with distal breakpoint beyond BGGP in ‘Red Egyptian’.
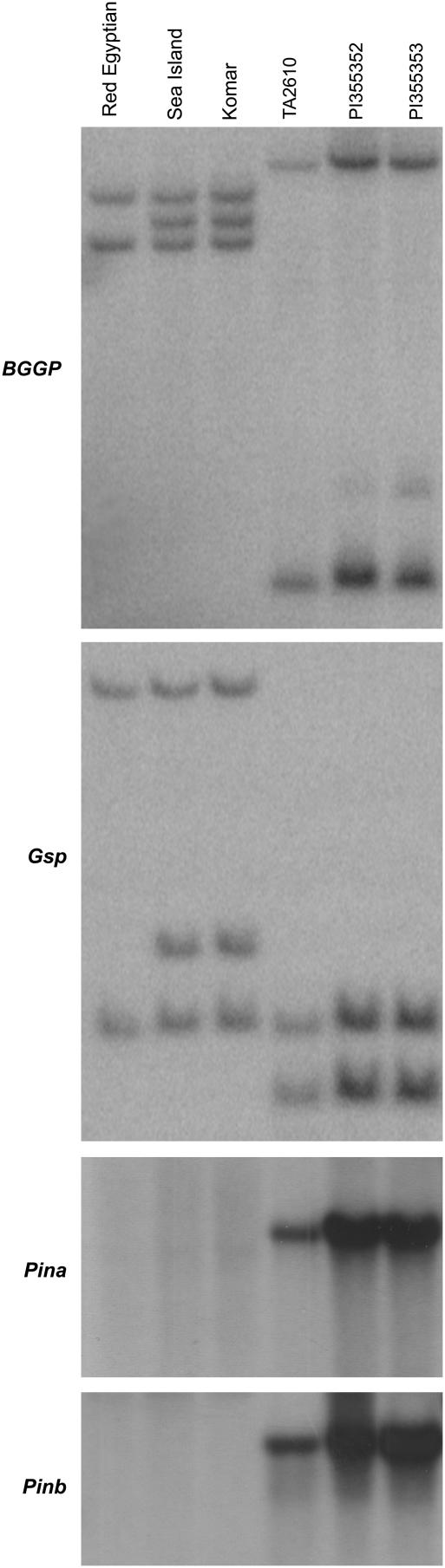
Figure 4.
Autoradiogram of Southern hybridization. Genes BBGP, Gsp, Pina, and Pinb as indicated at the left and accession numbers at the top. ‘Red Egyptian’, ‘Sea Island’, and ‘Komar’ are hard red cultivars of common wheat (T. aestivum); accessions TA2610, PI355352, and PI355353 belong to T. zhukovskyi. BGGP and Gsp each detected two fragments in ‘Red Egyptian’ and three in ‘Sea Island’ and ‘Komar’, whereas Pina and Pinb did not detect any signal in all three wheat cultivars. In T. zhukovskyi, BGGP and Gsp detected two bands with similar intensity and Pina and Pinb detected a single fragment.
We surveyed three accessions of T. zhukovskyi (AAmG), the second A-genome cluster hexaploid species. It is expected to have two copies of Pin genes, one from T. timopheevii and the second from T. monococcum. However, only one copy of the Pin genes was detected, indicating the presence of a second null haplotype at the Ha locus in one of its genomes (Fig. 4). Because T. zhukovskyi is autoallohexaploid, the loss of a Ha locus could be due to either recombination between A and Am genomes or a deletion event. This question can be resolved based on BGGP and Gsp hybridization patterns: (1) if each detects three bands, Pina and Pinb were deleted from one genome; (2) if two bands with similar intensity are observed, all BGGP, Gsp, Pina, and Pinb were deleted from the A or Am genome; and (3) if the two bands differ significantly in intensity, A-Am recombination, instead of deletion, occurred. Our results support the second scenario (Fig. 4); i.e. BGGP, Gsp, Pina, and Pinb were deleted from the A or Am genome, similar to the Ha-D haplotype in ‘Red Egyptian’.
We surveyed three D-genome cluster and one U-genome cluster hexaploid Aegilops spp. and all are expected to have two to three copies of Pina and three copies of Pinb, depending upon the genotype of the tetraploid parent (see above). A. crassa (DDX) and Aegilops vavilovii (Zhuk.) Chennav. (DXSs) were polymorphic; three accessions of A. crassa and two accessions of A. vavilovii had two copies of Pina and three copies of Pinb, and the rest had two copies of both Pina and Pinb. Aegilops juvenalis (Thell.) Eig (DXU) had three copies of Pina and two copies of Pinb in their genomes.
The U-genome cluster hexaploid A. neglecta (UnMnNn) was polymorphic; six accessions had two copies of Pina and three of Pinb, four accessions had one copy of Pina and three copies of Pinb, and the remaining three accessions had one copy of Pina and two copies of Pinb. Because tetraploid A. neglecta (UM) was polymorphic for Pina (one or two copies), the data suggest recurrent deletion of Pina at tetraploid and hexaploid levels in this species. Compared to Pina, Pinb deletion was only detected at the hexaploid level in the polyploid Aegilops spp.
Sequence Analyses of Unique Haplotypes
A sequence analysis of deletion haplotypes detected in polyploid wheat species was used to further characterize and allocate their genomic origin. These results are summarized in Figure 1.
The Ha-A Haplotype of T. timopheevii
Detection of the conserved Ha haplotype in the Timopheevi lineage of polyploid wheat triggered an immediate effort to determine its genomic origin. We designed A- and G-genome-specific primers based on Pina and Pinb sequences from diploid progenitors T. urartu and A. speltoides, respectively. A-genome-specific primers amplified strong fragments approximately 650 bp for Pina and 780 bp for Pinb. G-genome-specific primers produced very weak amplicons from T. timopheevii. Cloning and sequencing of the PCR products from six T. timopheevii accessions indicated that A-genome amplicons showed highest homology to Pin genes from T. urartu at the DNA and protein sequence levels, but G-genome amplicons showed no homology to the Pin genes, but weak similarities to various TEs. This clearly indicated that the A-genome haplotype (Pina-A and Pinb-A) was maintained and the G-genome copy was deleted in T. timopheevii.
The promoters and coding regions of Pina and Pinb are highly conserved among the timopheevii accessions and between timopheevii and urartu, except for a 1-bp insertion in the promoter region of Pina in one accession and an A-to-C transversion at position 181 of Pinb in another (Supplemental Figs. S1 and S2). In the 3′ region of Pinb, an 88-bp fragment, spanning the stop codon and polyadenylation signal, was found in triplicate in Pinb-A of T. timopheevii compared to its ancestor T. urartu. The repeat members are identical, except for a single-nucleotide polymorphism (Supplemental Fig. S2). A PCR assay showed that the 88-bp triple repeat is fixed at the species level (Fig. 5). A 7-bp sequence (CAACATG) was found at the beginning of each repeat member and immediately after the triple repeat in T. timopheevii and flanking the 88-bp sequence in T. urartu, suggesting that it originated by replication slippage after polyploidization (Supplemental Fig. S2). The alignments of nucleotide sequences of the Pina and Pinb genes and amino acid sequences of the PINA and PINB proteins between T. timopheevii and T. urartu are shown in Supplemental Figures S1, S2, S3, and S4.
Figure 5.
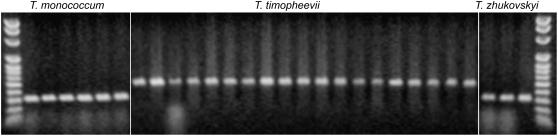
PCR assay of the 88-bp triple repeat at the 3′ end of Pinb-A. The 100-bp ladder is on either side and species are indicated at the top. Amplicons of T. urartu and T. zhukovskyi are 308 bp and those of T. timopheevii are 484 bp in length.
The Ha-G Haplotype of T. timopheevii
To determine the fine structure of Ha-G, particularly the deletion breakpoints, we isolated Ha-G from a fosmid library constructed from accession Tim01 of T. timopheevii. The Gsp-containing fosmid clone, 1E05, was sequenced at 8 times coverage and assembled into five contigs after prefinishing, totaling 41,262 bp. The largest contig, 23,807 bp, contains three non-TE protein-coding genes within 10,919 bp, BGGP, Gsp, and ATPase (Fig. 2). In addition, a 311-bp sequence, corresponding to the 5′ portion of Gene3 was found 1,806 bp downstream of Gsp and 2,749 bp upstream of ATPase, and a partial coding sequence of another copy of the ATPase gene was found 778 bp downstream of the intact ATPase at the 3′ end of the fosmid clone. At the nucleotide sequence level, Gsp-G is highly homologous to Gsp-S of A. speltoides and Gsp-B of T. turgidum and T. aestivum. ATPase-G showed highest identity (98%) to ATPase-1 (Gene7-1, pseudogene) from chromosome 5B of T. turgidum and T. aestivum. BGGP-G has high homology to BGGP-D of A. tauschii and T. aestivum and to BGGP-B of T. turgidum and T. aestivum. As expected, GSP-G is encoded by the plus strand, and BGGP-G and ATPase-G are encoded by the minus strand. The Ha-G haplotype lost Gene8 along with Pina, Pinb, ATPase-4, and ATPase-5, and has a distal breakpoint between Gene3 and Pina and a proximal breakpoint between Gene8 and ATPase-1. Compared to the Ha-S locus, more than 200 kb of sequence was deleted from the Ha-G and Ha-B loci during polyploid wheat evolution (Fig. 2).
The Ha-Am Haplotype of T. zhukovskyi
Because T. zhukovskyi originated from a cross between T. timopheevii (Ha-A, null Ha-G) and T. monococcum (Ha-Am), the conserved haplotype in this hexaploid wheat is either of Ha-A or Ha-Am origin. The Am genome of T. monococcum is highly homologous to the A genome of T. urartu due to their very recent divergence. Multiple sequence alignment of the Pin gene sequences available in the National Center for Biotechnology Information (NCBI) database from these two species and sequences of T. timopheevii from this research were used to identify species-specific single-nucleotide polymorphisms to unambiguously assign the genome origin of Pina and Pinb in T. zhukovskyi. A monococcum-specific synonymous point mutation was identified at position 384 within the open reading frame, where three accessions of T. monococcum carried an A, the mutant allele, and all accessions of T. urartu, T. timopheevii, and the remaining T. monococcum accessions carried a G, the wild-type allele (Supplemental Fig. S5). Similarly, a species-specific synonymous mutation was found at position 75 in the open reading frame of Pinb, where all the T. urartu and T. timopheevii accessions carry a T and all the accessions of T. monococcum carry a C (Supplemental Fig. S6). All three accessions of T. zhukovskyi carry monococcum-specific alleles, indicating that the Pina-A and Pinb-A from T. timopheevii were deleted and are survived by Pina-Am and Pinb-Am from T. monococcum (Supplemental Figs. S5 and S6). Furthermore, the 88-bp triple repeat was not detected in Pinb of T. zhukovskyi by either sequencing or PCR assay (Fig. 5), providing additional evidence that Pinb-A was deleted in this hexaploid wheat species.
The Ha Loci of Synthetic Tetraploids
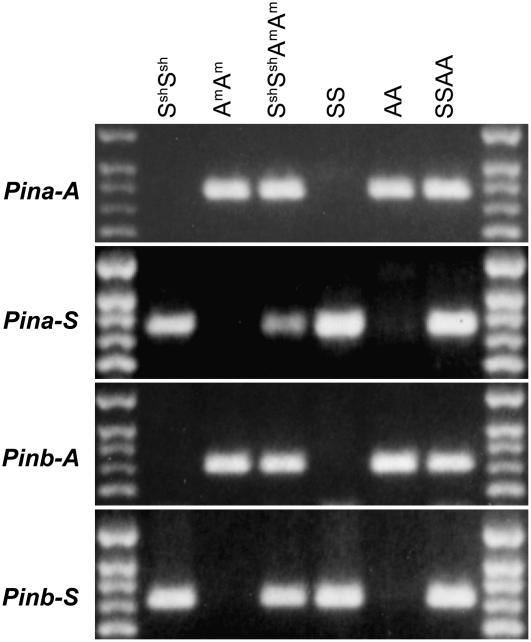
The fixation of Ha deletion patterns in tetraploid wheat suggested that Pin genes may have been eliminated immediately upon polyploidization similar to the low-copy sequences demonstrated by Feldman et al. (1997), Liu et al. (1998), and Ozkan et al. (2001). We tested this hypothesis on tetraploid AASS- and AmAmSshSsh-genome amphiploids. The AASS amphiploid was produced in our laboratory by crossing diploid species T. urartu (A) with A. speltoides (S), followed by chromosome doubling. The AmAmSshSsh was produced from T. monococcum (Am) and Aegilops sharonenesis Eig (Ssh) by Ozkan et al. (2001). We used genome-specific primers for both Pina and Pinb to produce amplicons for sequencing from the AASS and AmAmSshSsh amphiploids and their diploid parents. Pina and Pinb haplotypes were conserved in both amphiploids (Fig. 6). The Pina and Pinb sequences of the A and Am genomes between the amphiploids and their parents were identical (Supplemental Figs. S7, S8, S11, and S12), but up to 1% variation was found in the Pina and Pinb sequences of the S and Ssh genomes of the amphiploids compared to their Aegilops parents (Supplemental Figs. S9, S10, S13, and S14). More striking was a 21-bp deletion detected in Pinb-Ssh of the amphiploid AmAmSshSsh compared to its A. sharonensis parent. Six clones from that ligation were sequenced and the wild-type allele was not found. Pinb sequences of the S-genome species, including Aegilops bicornis (Forssk.) Jaub. and Spach (Sb), Aegilops longissima Schweinf. and Muschl. (Sl), Aegilops searsii Feldman and Kislev ex Hammer (SsSs), A. sharonensis (Ssh), and A. speltoides (S) contain two 11-bp direct repeats (GAAGTTGGCGG) separated by a 10-bp spacer (CTGGTACAAT). This 21-bp deletion was obviously caused by unequal crossing over between the 11-bp direct repeats (Supplemental Fig. S10) and led to a loss of seven amino acids (WYNEVGG) in the PINB protein. The AmAmSshSsh amphiploid used is from the S2 generation; the unequal crossover either occurred during female meiosis of the A. sharonensis parent TMB02 or happened and was rapidly fixed after polyploidization.
Figure 6.
PCR assay of Pina and Pinb in amphiploids and their parents. The 100-bp ladder is on either side and the lowest band is 500 bp. The genome formula is indicated at the top and gene symbols are at the left of the picture.
DISCUSSION
The most remarkable observation on the structure and evolution of the Ha locus in wheat and the Triticeae is the absolute conservation of the locus in diploid species reported here and in previous articles (Gautier et al., 2000; Lillemo et al., 2002; Massa et al., 2004; Chen et al., 2005; Simeone et al., 2006) and recurrent and independent deletions in the polyploid Triticum and Aegilops spp. To date, more than 200 accessions from the two diploid Triticum and 10 diploid Aegilops spp. have been analyzed and not a single case of deletion polymorphism at the Ha locus has been reported. Especially, no deletion polymorphisms have been detected in a diverse sample of more than 130 accessions of the A-, B-, and D-genome donor species of polyploid wheat. This is in contrast to frequent deletion haplotype polymorphisms for a defense-gene cluster in the D-genome diploid, A. tauschii (Brooks et al., 2006). Against this high rate of deletion polymorphism in polyploid species, not a single case of insertion-deletion polymorphism was documented in a sample of Pina and Pinb sequences from 50 accessions of diploid A. tauschii, representing its geographical diversity (Massa et al., 2004). All polyploid wheat and most polyploid Aegilops spp. harbored deletion haplotypes of independent origin at the Ha locus. So how does a gene that is essential in a diploid suddenly become deleterious so that it must be deleted in a polyploid? To begin to answer this question, some discussion about the nature of Pin genes, their function, the nature of gene action in polyploids and the mechanisms of polyploid genome evolution and speciation that promote expression and evolution of novel traits is needed.
Amino acid sequence analysis has shown that numerous storage proteins, including low-molecular-weight glutenin, α/β gliadins, lipid transfer proteins, trypsin inhibitor, α-amylase/trypsin inhibitor, GSP, PINA, and PINB, belong to the α-amylase inhibitors (AAIs) and seed storage (SS) protein subfamily because they have an AAI-SS domain. AAIs play an important role in the natural defense of plants against insects and pathogens mainly by inhibiting α-amylases and proteases. Puroindolines have bactericidal (Jing et al., 2003) and fungicidal activities (Krishnamurthy et al., 2001). PINA and PINB proteins directly bind to the surface of starch granules in the endosperm cells and form a friabilin complex. Using isogenic lines, Swan et al. (2006) showed that puroindolines seem to protect starch from microbial digestion and increased expression of PIN proteins decreased the starch digestibility of wheat in the rumen by up to 30%. α-Amylase is an important enzyme in starch metabolism and is induced in the aleurone by GA3 from the embryo during germination. Conceivably, wheat starch can be protected from α-amylase digestion by AAI activity of the PIN proteins. Therefore, because of the important role of PIN proteins in plant defense and seed physiology, Pina and Pinb genes may be under strong selection pressure and are maintained in all the diploid species.
One of the consequences of polyploidy is doubling and tripling of gene copy number and, thus, the amount of proteins may be doubled or tripled for some of these genes. This dosage response has been demonstrated for the Pin genes and a supersoft hexaploid wheat genotype has been created (See et al., 2004). Because wheat starch is protected from α-amylase digestion by AAI activity of the PIN proteins, we hypothesize that the sudden dosage-driven increase in expression levels of Pin genes in polyploid would impede the embryos from obtaining nutrition from the endosperm. The situation may be more severe when polyploid plants are under abiotic stress, such as heat and drought during grain filling, which adversely affects endosperm development. Point mutations in Pinb can liberate the PIN proteins from binding to the starch granule surface and cause significant difference in grain texture (Giroux and Morris, 1997, 1998; Morris et al., 2001); however, the PIN proteins with the AAI activity remain in the endosperm cells. Therefore, deletion of Pina-Pinb genes provides the most efficient mechanism to reduce AAI activity and is the least detrimental because they are structural genes. The reduction in PIN proteins, in turn, enhances the exposure of starch granules to α-amylases during germination to provide sufficient nutrition for new polyploid seedlings to compete in stand establishment with surrounding diploid populations. Thus, the deleterious action of a single gene may determine the fate of a new polyploid species and, therefore, we consider it a bottleneck speciation event.
To test our hypothesis of the deleterious effect of a high dose of Pin genes on seed physiology, a preliminary germination experiment was performed using seeds from hexaploid ‘Chinese Spring’ wheat plants with two (background control), four, and six doses of the Pina-Pinb genes (See et al., 2004). Seeds from each genotype were placed in petri dishes on wet filter paper at room temperature. After 24 h, the check cultivar ‘Chinese Spring’ (with two doses) sprouted and radicle and coleoptile were visible. The genotype with four doses showed sprouting activity, but the radicle and coleoptile were invisible. The genotype with six doses did not sprout until 48 h after imbibition and radicle and coleoptile were visible only after 96 h of imbibition. The negative correlation between the Pina-Pinb dosages and the rate of germination and growth was consistently observed in subsequent days. Although this experiment appears to demonstrate the deleterious effects of higher doses of Pin genes on seed germination and stand establishment, more rigorous experiments need to be conducted, including assaying AAI activity of PIN proteins and evaluating germination, growth, and vigor of seedlings of the isogenic lines with different doses of Pin genes under field conditions. But what are the chances of occurrence and fixation of such a rare deletion mutation in a small founder polyploid species population and what genes might compensate for this missing function?
Because at least one copy of the Pin genes is maintained in the vast majority of polyploid species, the loss and fixation of null haplotype copies of both Pin genes in tetraploid T. turgidum require explanation. As mentioned earlier, a number of proteins have AAI activity and higher doses of their expression may partially compensate for the loss of function of the Pin genes. Gsp was not involved in most deletion haplotype polymorphisms and may have some compensatory functional role in defending against microbe attacks. Because extensive resetting of gene expression patterns follows polyploidy, it is possible that other genes may have been recruited for the same role. In fact, quantitative trait loci for hardness phenotype have been mapped in other regions of the genome besides the 5DS locus (Breseghello et al., 2005; Narasimhamoorthy et al., 2006).
Accumulating evidence suggests that polyploidization is accompanied by significant genome restructuring and resetting of gene expression patterns (for review, see Chen and Ni, 2006; Chen, 2007). Numerous genetic and epigenetic changes have been observed in neopolyploids (amphiploids), including sequence elimination (Song et al., 1995; Feldman et al., 1997; Liu et al., 1998; Ozkan et al., 2001), chromosome rearrangements (Pontes et al., 2004), changes in methylation (Shaked et al., 2001) and gene expression patterns (Adams et al., 2003; He et al., 2003), reactivation of TEs (Kashkush et al., 2003), and microRNA expression (Tian et al., 2006). The rate of DNA sequence divergence is much higher in polyploid wheat compared to diploid relatives (for review, see Dubcovsky and Dvorak, 2007). The great abundance of repetitive sequences, especially retroelements, in the wheat genome (Li et al., 2004; Devos et al., 2005) seems to promote gene deletion/duplication events. Chantret et al. (2005) reported that the loss of a block of genes, including the Pina and Pinb genes from A and B genomes of tetraploid T. turgidum, occurred independently by illegitimate recombination among retroelements bordering the Ha locus. Such events appear to be common at the Ha locus because Chantret et al. (2005) documented two additional such events in polyploid wheat genotypes. The loss of a large gene block, including Ha-G in T. timopheevii as reported here, may have been caused by a similar mechanism. We also documented replication slippage as a mechanism leading to triple repeat of an 88-bp sequence at the Ha-A locus of T. timopheevii. Unequal crossing over led to the deletion of a 21-bp sequence in the Pinb-Ssh gene in the AmAmSshSsh synthetic amphiploid. Although Pina and Pinb were conserved in a number of tetraploid Aegilops spp., they may not be transcribed as documented by Chen et al. (2005). Others were polymorphic for the Pina and/or Pinb deletion haplotype and none of the hexaploid species tolerated three doses of Pin genes (Table I). Although there is a high rate of deletion in polyploid Triticum and Aegilops spp., polyploidy per se does not cause Pin gene deletions as indicated by our data on synthetic amphiploids. The high mutation rate at the Ha locus in polyploid species, coupled with high gametic transmission of deletion haplotypes due to polyploid buffering, and the high fitness cost of higher doses of Pin gene expression on seed physiology led to the fixation of deletion haplotypes at the Ha locus in the founder populations during polyploid wheat speciation.
Polyploidization leads to both additive and nonadditive gene expression patterns (for review, see Chen, 2007). Documentation of recurrent deletions at the Ha locus suggests certain rules for the fate of loci that show dosage-sensitive expression following polyploidization. If the additive gene expression has a deleterious effect on the organism, then it will be rapidly deleted. Conversely, if the additive gene expression has a beneficial effect, then it will be conserved. If the effect is neutral, then the duplicate loci may undergo subfunctionalization or mutation to assume new functions (Adams et al., 2003). Comparative genomics is generating large databases of gene duplications and deletions following whole-genome polyploidization. The challenge of the postgenomics era will be to determine the physiological bases of such duplication/deletion events.
MATERIALS AND METHODS
Plant Materials
Plant materials are summarized in Table I regarding the species, ploidy levels, genome formula, and number of accessions. The details of individual accessions are listed in Supplemental Table S1 with their Pina and Pinb scores. Accessions prefixed with TA are maintained by the Wheat Genetic and Genomic Resources Center, Kansas State University (Manhattan, KS); accessions prefixed with CItr, PI, and PVP were obtained from Dr. Harold Bockelman at the U.S. Department of Agriculture Small Grains Collection (Aberdeen, ID). Triticum timopheevii accession Tim01, the TH02/TMB02 amphiploid (AmAmSS), the Aegilops sharonensis (TH02), and Triticum monococcum (TMB02) were provided by Dr. Moshe Feldman, Weizmann Institute of Science (Rehovot, Israel). The amphiploid (TA3438, AASS) was derived from a cross made at the Wheat Genetic and Genomic Resources Center between Aegilops speltoides accession TA1785 and Triticum urartu accession TA831. All lines were grown in the greenhouse. The ploidy levels of Aegilops crassa and Aegilops neglecta accessions were determined by Badaeva et al. (1998, 2001) and this research. Critical accessions were identified by C banding (Gill et al., 1991) when necessary.
Clones and Primers
Wheat (Triticum aestivum) cDNA clones TMA9 (Pina) and TMA10 (Pinb) were provided by Dr. Marie-Françoise Gautier (Unité de Biochimie et Biologie Moléculaire des Céréales, Institut National de la Recherche Agronomique, France). EST BU100707 homologous to Gene1 was obtained from the Arizona Genomics Institute (Tucson, AZ). The BAC clone 197O23 of A. speltoides was provided by Dr. Jan Dvorak (University of California, Davis, CA). All PCR primers for amplification of Gsp, Pina, Gene5, and Pinb and annealing temperatures are given in Supplemental Table S2.
RFLP Analysis to Detect Deletions
The copy numbers of Pina and Pinb were determined based on the number of fragments detected in Southern blots. Approximately 100 mg of leaf tissue were collected from each accession, lyophilized in a 2-mL microcentrifuge tube, and disrupted by shaking with metal beads. The procedures for DNA extraction, digestion, electrophoresis, and Southern hybridization generally followed those of Faris et al. (2000). For most accessions, EcoRI, HindIII, or BamHI were used for digestion. The search of the coding sequences of Pina and Pinb deposited in the NCBI did not reveal restriction sites BamHI and EcoRI, and the HindIII restriction site was only found at position 24 of Pinb from the S-genome species. In some cases, more bands were observed than expected, possibly due to heterozygosity, intragenic restriction site, or duplication, and they were ignored because the focus of this work was on the detection of the Pin gene deletions.
Molecular Cloning
PCR products of genes Gsp, Pina, Gene5, and Pinb were separated by agarose gel electrophoresis, eluted from gel, ligated to T-easy vector (Promega), and transformed into Escherichia coli strain DH10B.
To clone the Ha locus from T. timopheevii, we constructed a fosmid library of accession Tim01. Briefly, total genomic DNA of Tim01 was sheared by 120 cycles of freezing in liquid nitrogen and thawing in a 65°C water bath, and separated by CHEF gel electrophoresis. Fragments of 30 to 50 kb were excised, eluted, end repaired with an End-It kit (Epicentre Biotech), and ligated to the CopyControl pCC1FOS. The ligation was packaged with MaxPlax Lambda Packaging Extracts (Epicentre Biotech), diluted 100-fold, and used to infect E. coli strain PE1300 following the manufacturer's instructions. An aliquot of 70 μL infected bacteria (approximately 70 clones) was distributed and maintained in 384-well plates. The library first was pooled by plate and screened by PCR using Gsp-S-specific primers. Positive plates were pooled by rows and columns. Once a positive well was identified, the culture from that well was spread onto Luria-Bertani agar plates containing chlorophenicol (12.5 μg/mL) and colonies were picked, arrayed in 96-well plates, and screened individually by PCR. To isolate the Ha locus from the G-genome donor species, we hybridized the Pina and Gsp to the macroarray filters of an A. speltoides BAC library (Akhunov et al., 2005).
Sequence Analysis
To design genome-specific primers, nucleotide sequences for Gsp, Pina, and Pinb of diploid species of Triticum and Aegilops were retrieved from NCBI (http://www.ncbi.nlm.nih.gov) and subjected to multiple sequence alignment with ClustalW software at Baylor College of Medicine (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) and formatted by BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). For the Pina and Pinb genes cloned from T. timopheevii, Triticum zhukovskyi, amphiploids, and their parents, eight clones from a ligation were sequenced, and the sequences were assembled with CAP3 program (Huang and Medan, 1999; http://pbil.univ-lyon1.fr/cap3.php), with all the parameters set to default. Sequence contigs were used as queries for BLASTn searches against the NCBI nonredundant database. A fosmid and a BAC were shotgun sequenced at 8 times equivalents and assembled with the program Consed (Gordon et al., 1998). Protein-coding genes were predicted using the program FGENESH (http://sun1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind) with the organism set as monocot. Repeated sequences were identified by searching the Triticeae Repeat Sequence database (Wicker et al., 2002) by BLASTn and BLASTp (http://wheat.pw.usda.gov/ITMI/Repeats/blastrepeats3.html).
Gene Nomenclature
Gene designations followed the rules of nomenclature as listed in the Wheat Gene Symbol Catalogue (McIntosh et al., 1998). The Ha locus is triplicated in common wheat and orthologous loci of A, B, and D genome origin are designated as Ha-A, Ha-B, and Ha-D, respectively. For hypothetical and function-unknown genes at the Ha locus, the names designated by Chantret et al. (2005) are adopted to avoid any confusion.
Sequence data for this article can be found in the GenBank/EMBL data libraries under accession numbers EU267678, EU267679, and EU268462 to EU268495.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of Pina sequences from the A genomes of wheat.
Supplemental Figure S2. Alignment of Pinb sequences from the A genomes of wheat.
Supplemental Figure S3. Alignment of PINA sequences from the A genomes of wheat.
Supplemental Figure S4. Alignment of PINB sequences from the A genomes of wheat.
Supplemental Figure S5. Alignment of Pina sequences from the A and Am genomes of wheat.
Supplemental Figure S6. Alignment of Pinb sequences from the A and Am genomes of wheat.
Supplemental Figure S7. Alignment of Pina-Am between the amphiploid and its parent.
Supplemental Figure S8. Alignment of Pina-Ssh between the amphiploid and its parent.
Supplemental Figure S9. Alignment of Pinb-Am between the amphiploid and its parent.
Supplemental Figure S10. Alignment of Pinb-Ssh between the amphiploid and its parent.
Supplemental Figure S11. Alignment of Pina-A between the amphiploid and its parent.
Supplemental Figure S12. Alignment of Pina-S between the amphiploid and its parent.
Supplemental Figure S13. Alignment of Pinb-A between the amphiploid and its parent.
Supplemental Figure S14. Alignment of Pinb-S between the amphiploid and its parent.
Supplemental Table S1. Plant materials and their scores on Pina and Pinb.
Supplemental Table S2. PCR primers for Gsp, Pina, Pinb, and Gene5.
Supplementary Material
Acknowledgments
We thank Dr. Harold Bockelman and Dr. Moshe Feldman for providing seeds, Dr. Jan Dvorak and Dr. Marie-Françoise Gautier for providing cDNA clones, Dr. Bernd Friebe for C-banding identification of wheat accessions, and Jon Raupp and Duane Wilson for technical assistance.
This work is contributed from the Kansas Agricultural Experiment Station (contribution no. 07–255–J) and was partially supported by grants from the U.S. Department of Agriculture and the Kansas Wheat Commission.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Bikram S. Gill (bsgill@ksu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov ED, Akhunova AR, Dvorak J (2005) BAC libraries of Triticum urartu, Aegilops speltoides and Ae. tauschii, the diploid ancestors of polyploid wheat. Theor Appl Genet 111 1617–1622 [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Amosova AV, Samatadze TE, Zoshchuk SA, Shostak NG, Chikida NN, Zelenin AV, Raupp WJ, Friebe B, Gill BS (2001) Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst Evol 246 45–76 [Google Scholar]
- Badaeva ED, Friebe B, Zoshchuk SA, Zelenin AV, Gill BS (1998) Molecular cytogenetic analysis of tetraploid and hexaploid Aegilops crassa. Chromosome Res 6 629–637 [DOI] [PubMed] [Google Scholar]
- Blochet JE, Chevalier C, Forest E, Pebay-Peyroula E, Gautier MF, Joudrier P, Pezolet M, Marion D (1993) Complete amino acid sequence of puroindoline, a new basic and cystine rich protein with a unique tryptophan-rich domain, isolated from wheat endosperm by Triton X-114 phase partitioning. FEBS Lett 329 336–340 [DOI] [PubMed] [Google Scholar]
- Blochet JE, Kabooulou A, Compoint JF, Marion D (1991) Amphiphilic proteins from wheat flour: specific extraction, structure and lipid binding properties. In W Bushuk, R Tkachuk, eds, Gluten Proteins 1990. American Association of Cereal Chemists, St. Paul, pp 314–325
- Breseghello F, Finneyc PL, Gainesd C, Andrewsd L, Tanakab J, Pennere G, Sorrells ME (2005) Genetic loci related to kernel quality differences between a soft and a hard wheat cultivar. Crop Sci 45 1685–1695 [Google Scholar]
- Brooks SA, Huang L, Herbel MN, Gill BS, Brown-Guedira G, Fellers JP (2006) Structural variation and evolution of a defense-gene cluster in natural populations of Aegilops tauschii. Theor Appl Genet 112 618–626 [DOI] [PubMed] [Google Scholar]
- Chantret N, Cenci A, Sabot F, Anderson O, Dubcovsky J (2004) Sequencing of the Triticum monococcum hardness locus reveals good microcolinearity with rice. Mol Genet Genomics 271 377–386 [DOI] [PubMed] [Google Scholar]
- Chantret N, Salse J, Sabot F, Rahman S, Bellec A, Laubin B, Dubois I, Dossat C, Sourdille P, Joudrier P, et al (2005) Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wilkinson M, Tosi P, He G, Shewry P (2005) Novel puroindoline and grain softness protein alleles in Aegilops species with the C, D, S, M and U genomes. Theor Appl Genet 111 1159–1166 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Ni Z (2006) Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Ma J, Pontaroli AC, Pratt LH, Bennetzen JL (2005) Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc Natl Acad Sci USA 102 19243–19248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J (1998) Genome analysis in the Triticum-Aegilops alliance. In AE Slinkard, ed, Proceedings of the 9th International Wheat Genetics Symposium. University Extension Press, Saskatoon, Canada, pp 8–11
- Dvorak J, di Terlizzi P, Zhang HB, Resta P (1993) The evolution of polyploid wheats: identification of A genome donor species. Genome 36 21–31 [DOI] [PubMed] [Google Scholar]
- Dvorak J, Yang ZL, You FM, Luo MC (2004) Deletion polymorphism in wheat chromosome regions with contrasting recombination rates. Genetics 168 1665–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris JD, Haen KM, Gill BS (2000) Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier MF, Aleman ME, Guirao A, Marion D, Joudrier P (1994) Triticum aestivum puroindolines, two basic cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol Biol 25 43–57 [DOI] [PubMed] [Google Scholar]
- Gautier MF, Cosson P, Guirao A, Alary R, Joudrier P (2000) Puroindoline genes are highly conserved in diploid ancestor wheats and related species but absent in tetraploid Triticum species. Plant Sci 153 81–91 [Google Scholar]
- Gill BS, Friebe B (2002) Cytogenetics, phylogeny and evolution of cultivated wheats. In S Rajaram, BC Curtis, H Gomez Macpherson, eds, Bread Wheat—Improvement and Production. FAO Plant Production and Protection Series No. 30. Food and Agriculture Organization, Rome, p 567
- Gill BS, Friebe B, Endo TR (1991) Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34 830–839 [Google Scholar]
- Giroux MJ, Morris CF (1997) A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor Appl Genet 95 857–864 [Google Scholar]
- Giroux MJ, Morris CF (1998) Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc Natl Acad Sci USA 95 6262–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8 195–202 [DOI] [PubMed] [Google Scholar]
- Greenwell P, Schofield JD (1986) A starch granule protein associated with endosperm softness in wheat. Cereal Chem 63 379–380 [Google Scholar]
- He P, Friebe BR, Gill BS, Zhou JM (2003) Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol Biol 52 401–414 [DOI] [PubMed] [Google Scholar]
- Huang S, Sirikhachornkit A, Su X, Faris J, Gill BS, Haselkorn R, Gornicki P (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA 99 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Medan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing W, Demeoe AR, Vogel HJ (2003) Conformation of a bactericidal domain of puroindoline a: structure and mechanism of action of a 13-residue antimicrobial peptide. J Bacteriol 185 4938–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33 102–106 [DOI] [PubMed] [Google Scholar]
- Kihara H (1944) Die entdeckung des DD-analysators beim. Weizen Agric Hortic 19 889–890 [Google Scholar]
- Kilian B, Ozkan H, Deusch O, Effgen S, Brandolini A, Kohl J, Martin W, Salamini F (2007) Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol 24 217–227 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy K, Balconi C, Sherwood JE, Giroux MJ (2001) Wheat puroindoline enhance fungal disease resistance in transgenic rice. Mol Plant Microbe Interact 14 1255–1260 [DOI] [PubMed] [Google Scholar]
- Law CN, Young CF, Brown JWS, Snape JW, Worland AJ (1978) The study of grain protein control in wheat using whole chromosome substitution lines. In Seed Protein Improvement by Nuclear Techniques. International Atomic Energy Agency, Vienna, pp 483–502
- Li W, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40 500–511 [DOI] [PubMed] [Google Scholar]
- Lillemo M, Simeone MC, Morris CF (2002) Analysis of puroindoline a and b sequences from Triticum aestivum cv. “Penawawa” and related diploid taxa. Euphytica 126 321–331 [Google Scholar]
- Liu B, Vega JM, Feldman M (1998) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41 535–542 [DOI] [PubMed] [Google Scholar]
- Massa AN, Morris CF, Gill BS (2004) Sequence diversity of puroindoline-a, puroindoline-b, and the grain softness protein genes in Aegilops tauschii Coss. Crop Sci 44 1808–1816 [Google Scholar]
- Mattern JM, Morris R, Schmidt JW, Johnson VA (1973) Location of genes for kernel properties in wheat cultivar “Cheyenne” using chromosome substitution lines. In ER Sears, LMS Sears, eds, Proceedings of the 4th International Wheat Genetic Symposium. University of Missouri, Columbia, MO, pp 703–707
- McFadden ES, Sears ER (1946) The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered 37: 81–89, 107–116 [DOI] [PubMed]
- McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ (1998) Catalogue of gene symbols for wheat. In AE Slinkard, ed, Proceedings of the 9th International Wheat Genetics Symposium. University Extension Press, Saskatoon, Canada, pp 1–235
- Morris CF (2002) Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol 48 633–647 [DOI] [PubMed] [Google Scholar]
- Morris CF, Lillemo M, Simeone MC, Giroux MJ, Babb SL, Kidwell KK (2001) Prevalence of puroindoline grain hardness genotypes among historically significant North American spring and winter wheats. Crop Sci 41 218–228 [Google Scholar]
- Narasimhamoorthy B, Gill BS, Fritz AK, Nelson JC, Brown-Guedira GL (2006) Advanced backcross QTL analysis of a hard winter wheat × synthetic wheat population. Theor Appl Genet 112 787–796 [DOI] [PubMed] [Google Scholar]
- Nesbitt M, Samuel D (1996) From staple crop to extinction? The archaeology and history of hulled wheats. In S Padulosi, K Hammer, J Heller, C Pacoli, eds, Hulled Wheats. Promoting the Conservation and Use of Underutilized and Neglected Xrops. Proceedings of the 1st International Workshop on Hulled Wheats. International Plant Genetic Resources Institute, Rome, pp 41–100
- Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS (2004) Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci USA 101 18240–18245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See DR, Gioroux M, Gill BS (2004) Effect of multiple copies of puroindoline gene on grain softness. Crop Sci 44 1248–1253 [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone M, Gedye KR, Mason-Gamer R, Gill BS, Morris CF (2006) Conserved regulator elements identified from a comparative puroindoline gene sequence survey of Triticum and Aegilops diploid taxa. J Cereal Sci 44 21–33 [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92 7719–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan CG, Bowman JGP, Martin JM, Giroux MJ (2006) Increased puroindoline levels slow ruminal digestion of wheat (Triticum aestivum L.) starch by cattle. J Anim Sci 84 641–650 [DOI] [PubMed] [Google Scholar]
- Tian L, Chen M, Wang J, Wei NE, Sze SH, Chen ZJ (2006) The role of microRNAs in non-additive gene regulation in Arabidopsis allopolyploids. Plant & Animal Genomes XIV Conference. http://www.intl-pag.org/14/abstracts/PAG14_W363.html
- Tsunewaki K (1993) Genome-plasmon interaction in wheat. Jpn J Genet 68 1–34 [Google Scholar]
- Upadhya MD, Swaminathan MS (1963) Genome analysis in Triticum zhukovskyi, a new hexaploid wheat. Chromosoma 14 589–600 [Google Scholar]
- Wicker T, Matthews DE, Keller B (2002) TREP: a database for Triticeae repetitive elements. Trends Plant Sci 7 561–562 [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B (2001) Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J 26 307–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.