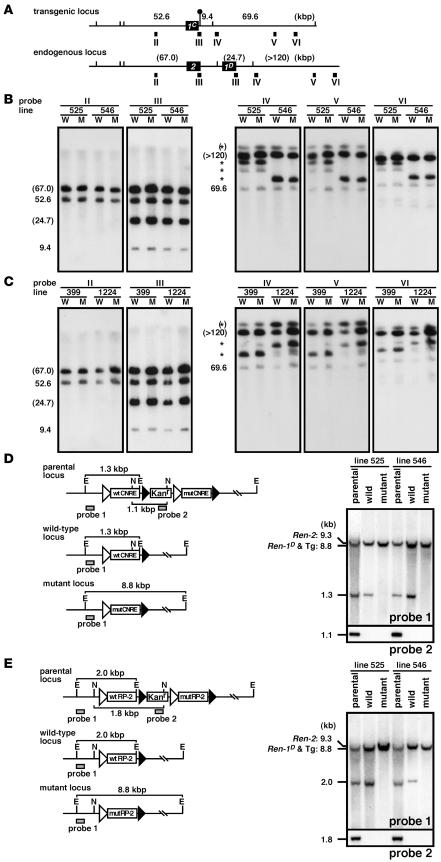
Figure 2. Structural analysis of the Tg lines.
(A) Comparison of the Tg Ren-1C and endogenous Ren-2 and Ren-1D loci. The renin genes and SfiI sites are shown as filled boxes and vertical lines, respectively. The artificially introduced SfiI site is marked by a lollipop. Expected fragment sizes (in kbp) after complete enzyme digestion are shown above the line. The probes used for Southern blot analysis in B and C are indicated by solid rectangles (II–VI). (B and C) Southern blot analyses of the TgM carrying either CNRE (B) or RP-2 (C) modifications. Thymic cells from WT (W) and mut (M) TgM lines were embedded in agarose plugs, digested with SfiI, and separated by PFG electrophoresis. DNA blots were hybridized separately to probes (II–VI) shown in A. The sizes of the expected bands are indicated (in kbp) on the left of each panel. Those expected from the endogenous Ren2 and Ren-1D loci are in parentheses, and partially digested DNA is marked by asterisks. (D and E) Fine structural analyses of the CNRE (D) and RP-2 (E) promoter regions. (Left panels) In vivo Cre/loxP recombination removes the Kanr gene plus either mut or WT promoter sequences from the parental locus to generate either the WT or the mut locus, respectively. E, EcoRI; N, NcoI. (Right panels) Tail DNA from parental, WT, and mut TgM was digested with EcoRI (top) or NcoI (bottom), separated on agarose gels, transferred to a nylon membrane, and hybridized with probes 1 and 2 (shaded rectangles in the left panels), respectively.