Fig. (3). Contrasting Degrees of Sequence Conservation of Ligand-binding Residues in VDR, PXR, and CAR.
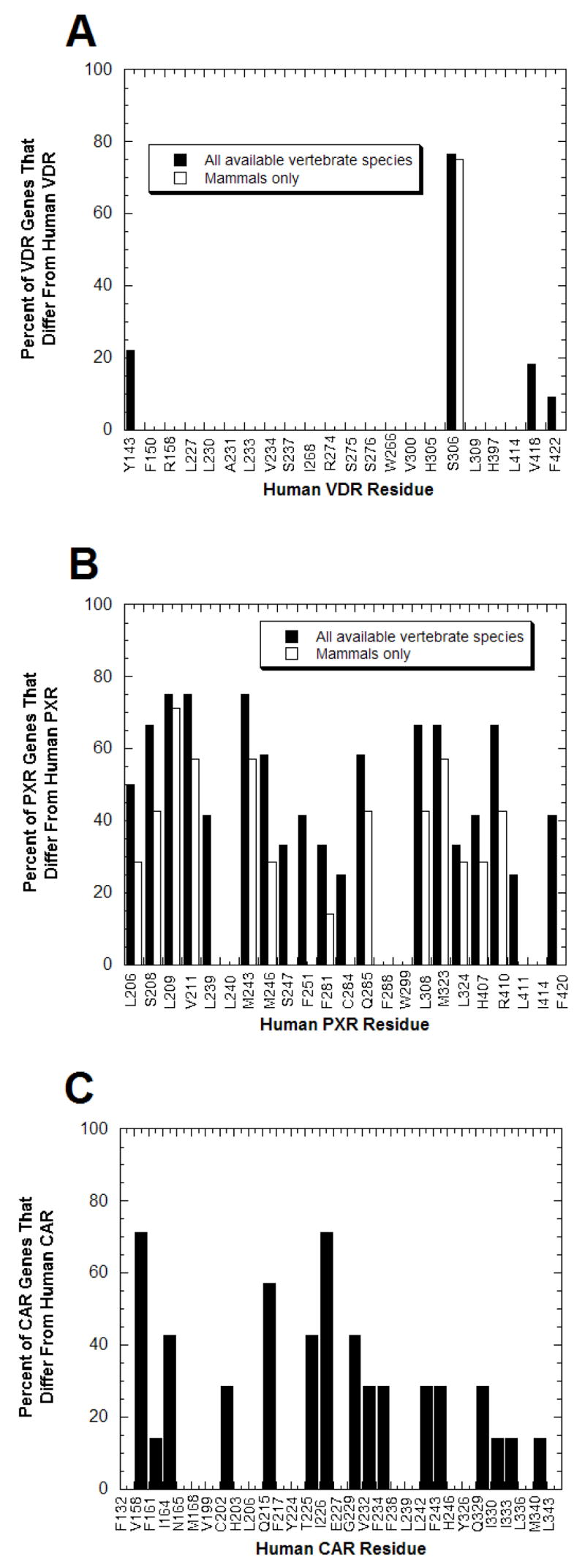
From the x-ray crystallographic structures of VDRs, PXRs, and CARs described in the legend to Fig. (2), the amino acid residues that directly interact with ligands are known. (A) Of the 22 amino acid residues shown to interact with ligands at human and rat VDRs, only 4 residues show any sequence variation across vertebrate species; the remaining 18 residues are completely conserved across all species. Due to partial sequence, data was missing for the four most C-terminal ligand-binding residues (corresponding to human VDR residues His397, Leu414, Val418, and F422) for crocodile, frog, fugu-β, lizard, snake, and turtle VDRs; data for the chimpanzee VDR was only available for the first two ligand-binding residues (corresponding to human VDR residues Tyr142 and Phe150). (B) PXRs contrast with VDRs in showing extensive amino acid sequence divergence at the positions corresponding to human PXR ligand-binding residues. Only 3 of 23 positions are conserved throughout the 13 vertebrate PXRs while for 9 of 23 residues, over half of the PXRs differ from the human PXR residue. (C) CARs also show significant sequence divergence at ligand-binding residues but not to the same magnitude as PXRs. The data is based on eight mammalian CAR sequences.
