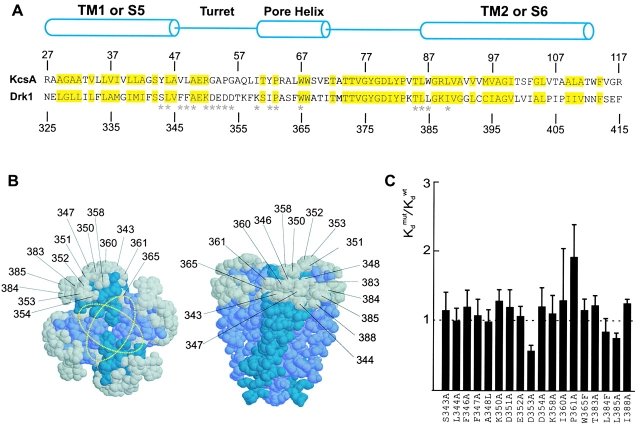
Figure 7.
Effects of the mutations in the peripheral surface of the pore domain. (A) Sequence alignment for the pore domain of the drk1 K+ channel and transmembrane part of KcsA. Numbers above and below the sequences indicate the positions of the residues for each channel. Yellow highlighting marks conserved residues between the two K+ channels. Cylinders indicate the demarcations of the two transmembrane α helices (TM1 and TM2 for KcsA and S5 and S6 for the drk1 K+ channel) and the pore helix. *18 positions in the drk1 K+ channel where mutations were made and the effects on Hanatoxin binding affinity were determined. All residues were mutated to Ala except A348L, W365F, and L384F. (B) Location of KcsA residues (shown in gray) that correspond to those mutated in the drk1 K+ channel. Gray residues correspond to those marked by asterisks in A. The crystal structure of the tetrameric KcsA channel shown for a top view (left) or a side view (right). Numbers correspond to residues in the drk1 K+ channel. Yellow dotted rings show two orientations of the Agitoxin2 footprint. (C) Normalized K d values for 18 mutations in the pore domain of the drk1 K+ channel. Data are mean ± SEM with n = 3 or 4 for each mutant channel. Dashed line marks a normalized K d value of 1.