Figure 8.
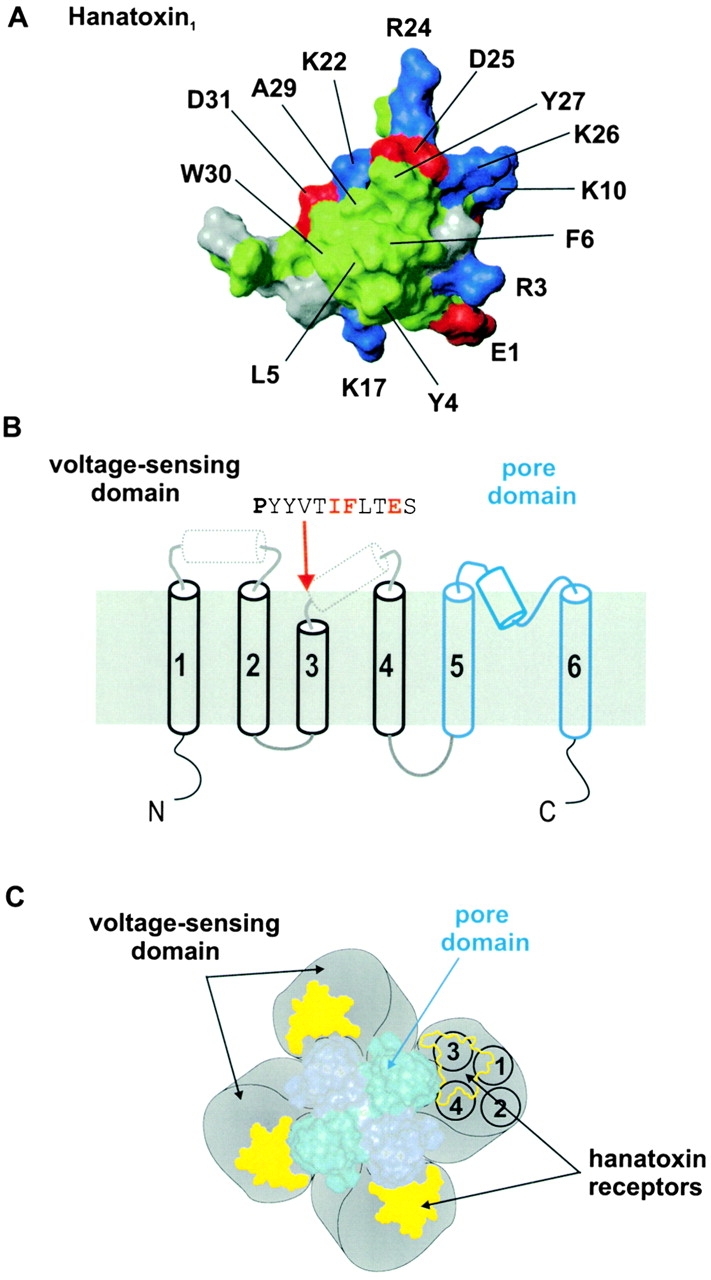
Molecular determinants and localization of the Hanatoxin receptors on the drk1 K + channel. (A) Surface rendering for one face of Hanatoxin1. The structure is a solution structure solved by NMR spectroscopy (Takahashi et al. 2000). The cluster of hydrophobic residues (green) surrounded by basic (blue) and acid (red) residues represents a surface feature common to Hanatoxin1, CsEV (an α-scorpion toxin) and ATXIII (a sea anemone toxin). (B) Putative topology and secondary structure of voltage-gated K+ channel α subunits. The first four transmembrane segments (S1–S4, in black) comprise the voltage-sensing domain and the last two transmembrane segments (S5–S6, in blue) in the pore domain. See Fig. 1 legend for details of secondary structure. The top is extracellular and the bottom is intracellular. A short stretch of sequence is shown for the COOH-terminal part of S3, beginning with the P268 (bold), a residue that is conserved in voltage-gated K+ channels. Red lettering indicates the positions of the three residues (I273, F274, and E277) that interact intimately with Hanatoxin. (C) Architecture of a voltage-gated K+ channel containing a central pore domain and four surrounding voltage-sensing domains drawn according to Li-Smerin et al. 2000b. Black circles shown for one voltage-sensing domain illustrate the four transmembrane (S1–S4) helices contained within, positioned according to Li-Smerin et al. 2000a. Yellow patches illustrate the Hanatoxin receptors confined within the voltage-sensing domains.
