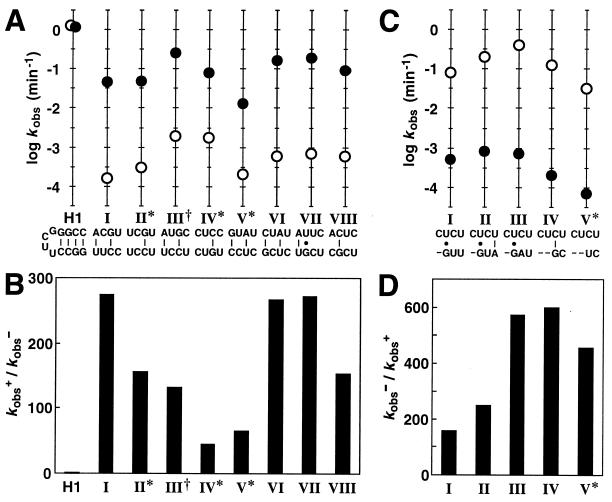
Figure 2.
Bridge sequences and kinetic parameters for individual allosteric ribozymes. (A) Sequences and corresponding ribozyme rate constants for eight classes of induction elements isolated from G6. Plotted for each class is the logarithm of the observed rate constant for self-cleavage in the absence (○) or presence (●) of FMN. The base-pairing schemes depicted for each bridge were generated by assuming that no base-pair shift relative to the G-C base pair remaining in stem II had occurred. Indicated are classes that display greater than 20% misfolding (∗) and a class wherein an extraneous mutation exists in the stem-loop region of the aptamer domain (†). H1 is an unmodified hammerhead ribozyme (5, 8) that displays maximum catalytic activity and that remains unaffected by the presence of FMN. (B) Fold-activation of catalytic activity (kobs+/kobs−) achieved in the presence of ligand for each class of FMN-inducible ribozyme. (C) Sequences and corresponding ribozyme rate constants for five classes of inhibition elements isolated from G6. Nucleotide deletions are represented as dashes. (D) Fold-inhibition of catalytic activity (kobs−/kobs+) achieved in the presence of ligand for each class of FMN-inhibited ribozyme.