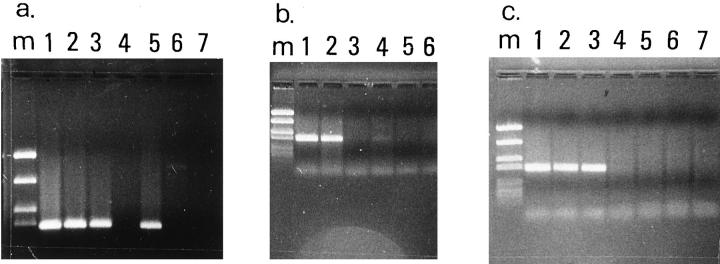
Figure 9.
Representative results of the PCR-based methylation assay. a: De novo methylation of CDKN2/p16INK4a exon 1a in a undifferentiated NSCLC (case 4). Lane m, marker (PUC19/Sau3AI); lane 1, undigested PCR product from HeLa cell line included as control; lanes 2 and 3, undigested PCR products from matched normal-tumor sample; lanes 4 and 5, digestion with methylation-sensitive enzyme HpaII revealed that CDKN2/p16ink4a in nontumor tissue is unmethylated as no amplification by PCR is seen, whereas in the tumor, CDKN2/p16ink4a is methylated as digestion does not affect amplification; lanes 6 and 7, digestion with non-methylation-sensitive enzyme MspI affects amplification in normal and tumor tissue. b: De novo methylation of CDKN2/p16ink4a exon 1a in a squamous cell lung carcinoma (case 11). Lane m, marker (PUC19/Sau3AI); lanes 1 and 2, undigested PCR products from matched normal-tumor sample; lanes 3 and 4, digestion with methylation-sensitive enzyme KspI revealed that CDKN2/p16ink4a in nontumor tissue is unmethylated as no amplification by PCR is seen, whereas in the tumor, CDKN2/p16ink4a is methylated as digestion does not affect amplification; lanes 5 and 6, digestion with non-methylation-sensitive enzyme MspI affects amplification in normal and tumor tissue. c: An adenocarcinoma with no indication of CDKN2/p16ink4a methylation (case 25). Lane m, marker (PUC19/Sau3AI); lane 1, undigested PCR product from HeLa cell line; lanes 2 and 3, undigested PCR products from the sample; lanes 4 and 5, digestion with methylation-sensitive enzyme KspI; lanes 6 and 7, digestion with non-methylation-sensitive enzyme MspI.