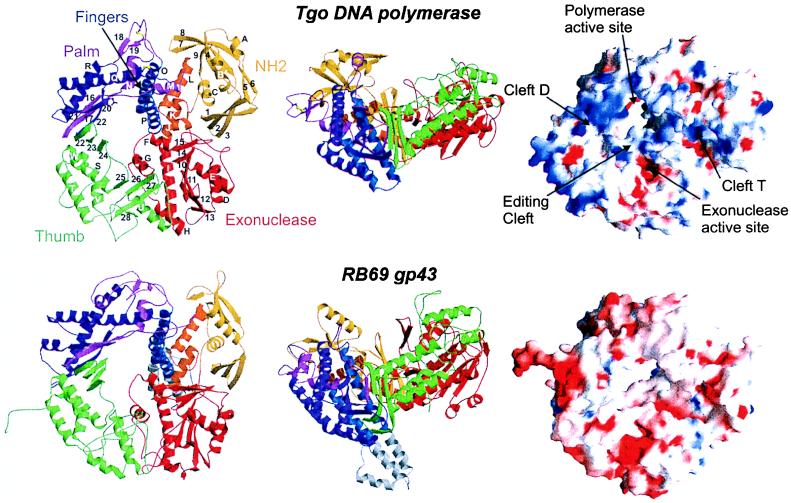
Figure 2.
Structure of Tgo pol and comparison with gp43 form bacteriophage RB69. (Left and middle) Ribbon representation of Tgo DNA polymerase (Upper) in two orthogonal views with labeled secondary structure elements. The molecule is composed of five domains: N-terminal domain (yellow), 3′ → 5′ exonuclease (red), palm [light and dark magenta represent the N-terminal and C-terminal segment, respectively (see text)], fingers (blue), and thumb (green), which are arranged to form a ring. An interdomain helical segment between the exonuclease domain and the palm is orange. The conserved carboxylates in the active site and the two disulfide bridges are shown as magenta and yellow ball-and-sticks, respectively. Tgo pol has the same overall architecture and domain topology than gp43 of RB69 (Lower). The 50-residue insertion in the fingers of gp43 is gray. (Right) Comparison of molecular surfaces of Tgo pol (Upper) and RB69 gp43 (Lower). Red and blue denote negative and positive electrostatic surface potentials, respectively. In contrast to gp43, Tgo pol has a strongly enhanced positive potential at the putative DNA-binding clefts.