Figure 4.
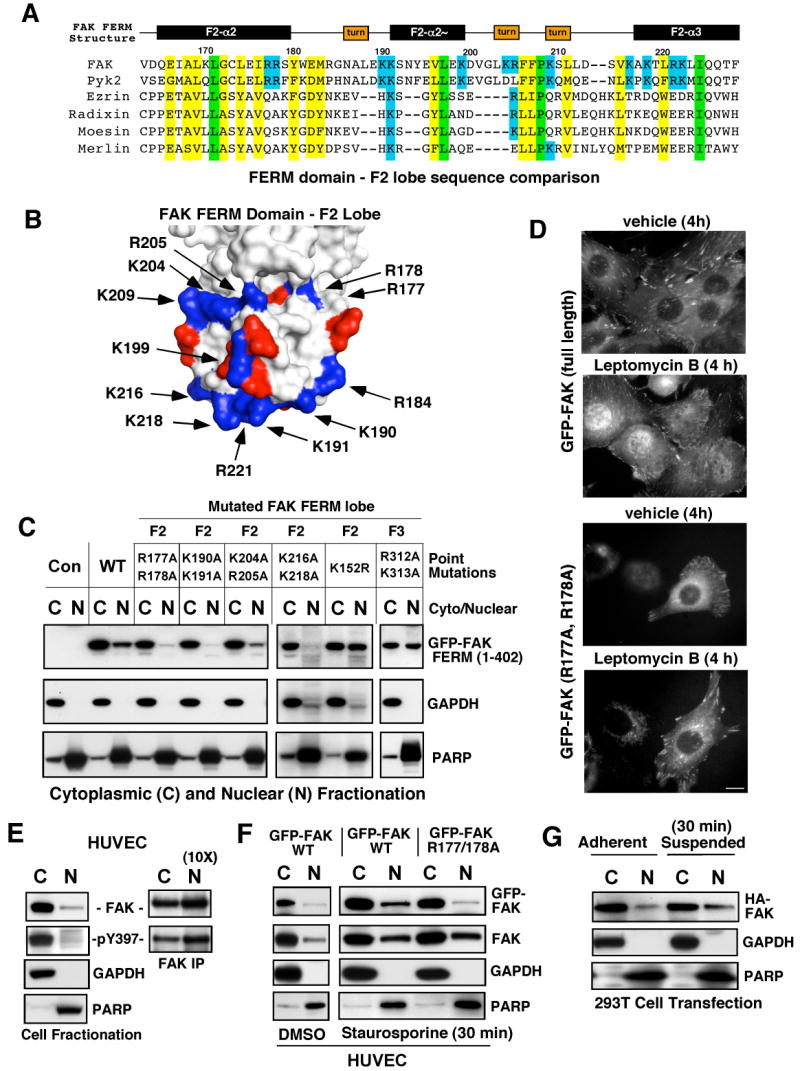
Determinants of FAK nuclear localization. (A) Structure-based alignment of FAK FERM F2 lobe residues (Lietha et al., 2007) with other FERM-containing proteins. Conserved basic residues within FAK and Pyk2 are highlighted in blue, total conserved FERM residues are highlighted in yellow and identical residues in green. (B) Localization of basic residue clusters on the surface of the FAK FERM F2 lobe. The FAK FERM domain (Lietha et al., 2007) F2 lobe was visualized using MacPyMOL. Basic residues (blue) are numbered according to the primary FAK sequence and acidic residues (red) are indicated. A putative nuclear targeting motif is comprised of residues at the tip of the F2 lobe (K190, K191, K216, K218, and R221). (C) FAK FERM domain analyzed by cellular fractionation. The indicated residues within GFP-FAK FERM (1-402) were mutated and constructs expressed in 293T cells. Cell lysates were separated into cytosolic (C) and nuclear (N) fractions, resolved by SDS-PAGE, and anti-GFP blotting was used to detect FAK FERM. Antibodies to gylceraldehyde-3-phosphate dehydrogenase (GAPDH) and poly ADP-ribose polymerase (PARP) were used to verify fractionation specificity, respectively. (D) Live cell imaging was used to follow GFP-FAK WT and GFP-FAK R177/178A distribution upon leptomycin B (10 ng/ml) or ethanol (vehicle) addition for 4 h. Scale bar is 10 μm. (E) FAK is partially nuclear-localized. HUVECs were separated into cytosolic and nuclear fractions, and blotted for FAK, GAPDH, and PARP. Ten-fold excess nuclear lysates was used to analyze FAK tyrosine phosphorylation by IP. (F) WT but not R177/178A FAK nuclear accumulation by HUVEC fractionation. HUVECs were treated with 1 μM staurosporine for 30 min, lysates separated into cytosolic or nuclear fractions, and immunoblotted with antibodies to GFP, FAK, GAPDH, and PARP. (G) 293T cells were transfected with HA-FAK and then fractionated into cytosolic and nuclear fractions under adherent and suspended conditions. Lysates blotted with anti-HA, GAPDH, and PARP.
