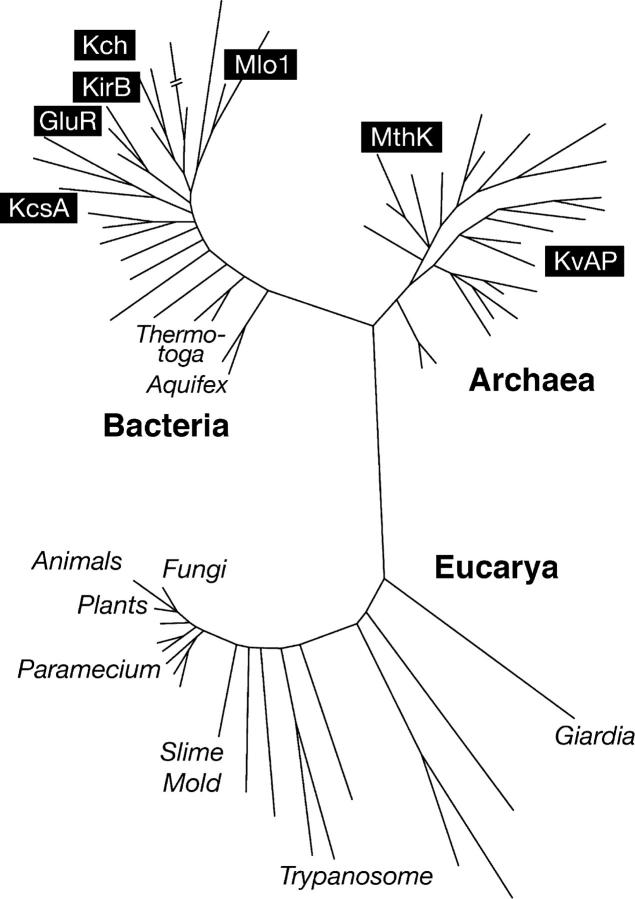
Figure 1.
A universal phylogenetic tree based on ssrRNA sequences, showing the three domains of life. Note that animals, plants and fungi constitute only a small portion of biological diversity, even among the eukaryotes. Some of the organisms mentioned in the text are marked. Marked are also the K+ channels, the crystal structures of which have been fully or partially solved: KvAP (6TM, voltage sensitive) from Aeropyrum pernix, a thermophilic archaeon, member of Crenarchaeota; MthK (2TM + RCK, Ca2+ binding, with a gating ring) from the methanogen Methanobacterium thermoautotrophicum, a member of Euarchaeota; Mlo1 (6TM + cyclic nucleotide–binding domain) from the soil bacterium Mesorhizobium loti, an α-proteobacterium (Gram negative); Kch (6TM + RCK) from Escherichia coli, a γ-proteobacterium; KirB (KirBAC1.1) a 2TM inward rectifier from Burkolderia pseudomalli, a rice pathogen of the β proteobacterium subdivision; GluR (GluR0) from the photosynthetic bacterium Synechocystis sp (cyanobacterium); KcsA (2TM) from Steptomyces lividans, an actinobactium (Gram positive). Lengths of the branches indicate differences in the nucleotide sequence of the small subunit ribosomal RNAs as a measure of the relatedness of different organisms. Modified from Pace (1997).