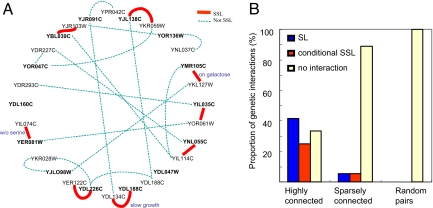
Fig. 3.
Results of the synthetic sick and lethal double-knockout experiments. (A) Pairs of dispensable genes for which genetic interaction was tested are connected by a solid red line in cases where SSL interaction was found and by a dashed blue line in cases where no interaction was observed. The hub-paralog pairs are arranged clockwise, starting from 12:00 (hub YJL138C, followed by its paralog YKR059W); all hubs are designated in boldface type. As a negative control, we codeleted hubs and randomly picked paralogs of other hubs. In instances of double knockout of the following hubs (YER081W and YMR105C) and their respective paralogs (YIL074C and YKL127W, respectively), SSL interactions were obtained only in specific growth conditions (lack of serine and galactose as a carbon source, respectively). Four hubs (YER081W, YDL226C, YJL098W, and YOR136W) were found to have two or three paralogs. For these cases, we searched for SSL interactions with all paralogs, yet we never found additional interactions (data not shown). For two hubs, we were unable to examine genetic interactions, either because of the essential nature of the hub itself (YDL047W, which in the database appears as viable, yet in our experiments, with specific genetic background, is extremely sick) or because of very low spore viability (YDL160C). (B) Proportions of the different genetic interactions obtained in all three double-knockout experiments are shown. Highly connected, double-knockout experiments in which both the highly connected gene and its duplicate were deleted; sparsely connected, double-knockout experiments in which both the sparsely connected gene and its duplicate were deleted; random pairs, double-knockout experiments in which both the highly connected gene and a randomly chosen paralog of another hub were deleted; SL, synthetic lethality; conditional SSL, lethality under specific conditions and slow growth; no interaction, no detectable fitness effect under the conditions tested.