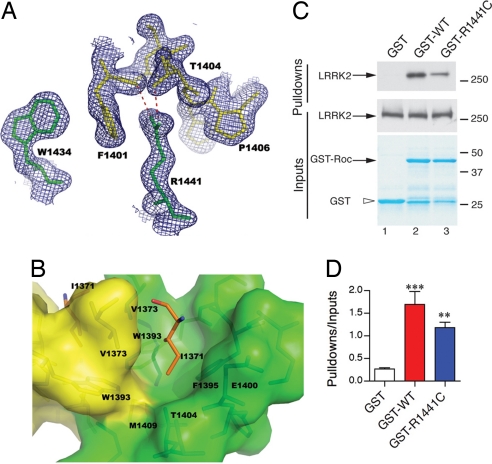
Fig. 2.
Structural basis of PD-associated mutations in ROC. (A) R1441 and W1434 from one monomer together with F1401 and P1406 from the other stack on each other alternately, forming a hydrophobic zipper at the dimer interface. The guanidinium group of R1441 also is hydrogen-bonded with the backbone carbonyl oxygen of F1401 and the hydroxyl group of T1404 on helix α2 from the other peptide chain. 2mFo − DFc electron density map is shown in blue. (B) I1371 is inserted in a hydrophobic cavity, which is constructed by residues from both monomers at the dimer interface. I1371 is shown in stick format and colored in orange. The surrounding residues are shown in stick format within the semitransparent surface representation. The color scheme is the same as that in Fig. 1. Note the side-chain methyl group of T1404 is pointing directly to the tip of I1371, forming a favorable van der Waals' interaction. (C) R1441C (lane 3), as a prototypical mutation at the dimer interface, decreases interaction with the full-length wild-type LRRK2 protein compared with wild-type GST fusions (lane 2); no interaction was seen with GST alone (lane 1). (D) Pull-down assays were quantified and corrected for the amount of LRRK2 protein in the inputs (middle blots). *, P < 0.0001; **, P < 0.01 compared with GST alone (one-way ANOVA with Student–Newman–Kuell's post hoc test; n = 3).