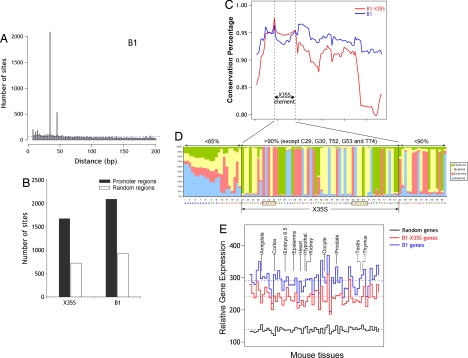
Fig. 2.
Comparison between B1-X35S and consensus B1. (A) Scan of the mouse proximal upstream genomic database for XRE and the nucleotide sequence that substitutes the Slug binding site in consensus B1 (CAGGCG). A set of >2,000 sites contained XRE and CAGGCG within 35 bp of each other. Theoretical distribution of 5-mer and 6-mer sites in a random genome would produce the result shown by the dashed blue line. (B) Differential location of B1-X35S and B1 in promoter or other regions in the mouse genome. Scanning was performed as indicated in Materials and Methods by using 23,459 random regions of 5 kbp in length. (C) Conservation analysis of B1-X35S (red) and B1 (blue) retrotransposons. Each value represents the mean conservation obtained for a window of 20 nucleotides. A set of 500 sequences was used for each B1 subfamily. The X35S region is indicated. (D) The nucleotide position-specific weight matrix of 1,673 X35S sites. Inside the X35S, nucleotide homology exceeds 90%, whereas outside of the X35S region (black vertical lines), the sequence conservation decreases. The consensus sequence is shown at the bottom, with the XRE and Slug sites boxed. (E) Comparative gene expression analysis for genes lacking the XRE and the Slug-binding site (random, black), containing B1-X35S (red), or B1 (blue). Each set included 800 genes whose expression was analyzed in a panel of 61 mouse tissues from the Gene Expression Atlas. The data are shown as the median for each tissue. The horizontal dashed lines represent the mean of all of the tissues. The data were normalized by the robust multichip average method. Differences in the medians were statistically significant, as assessed by paired t test (random vs. B1-X35S, random vs. B1, and B1-X35S vs. B1; P < 2.2 × 10−16). The Mann–Whitney statistical analysis for the mean B1-X35S vs. B1 gene expression was significant for the mouse tissues indicated (P < 0.05).