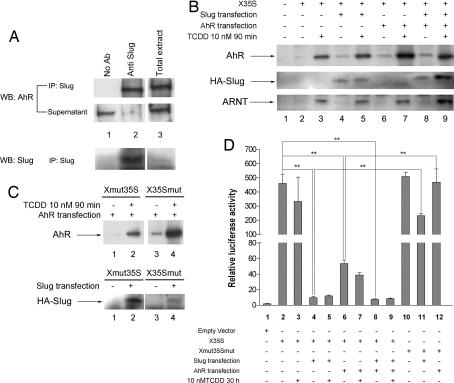
Fig. 3.
Functional analyses of X35S in vitro. (A) Nuclear extracts from basal Hepa 1 cells were immunoprecipitated with the anti-Slug antibody, and the AhR protein was detected by Western blotting. The total extract was used as positive control. The efficiency of the immunoprecipitation was determined by analyzing the protein remaining in the supernatants. (B) The DNA-binding affinity for AhR and HA–Slug on X35S. WT X35S was end-labeled with biotin and coupled to streptavidin paramagnetic particles. Nuclear extracts from untreated (DMSO) or TCDD-treated (10 nM for 90 min) Hepa 1 cells were incubated with X35S. AhR and HA–Slug binding was determined by Western immunoblotting by using antibodies specific for each protein. Where indicated, Hepa 1 cells were transfected with HA–Slug, AhR, or both expression vectors. The binding of the AhR dimerization partner ARNT was also determined under the same experimental conditions. (C) The DNA-binding affinity of AhR and HA–Slug to the mutated XRE (Xmut35S) or Slug site (X35Smut). Experimental conditions were as indicated above. (D) Luciferase reporter assays were used to analyze X35S activity in vitro. Hepa 1 cells were left untreated (DMSO) or treated with 10 nM TCDD for 24 h. Where indicated, cultures were transfected with HA–Slug, AhR, or both. Firefly luciferase activity was normalized by renilla luciferase (relative luciferase activity). The data shown are means ± SE from three experiments in triplicate. Differences among experimental conditions are significant at P = 0.001 (**).