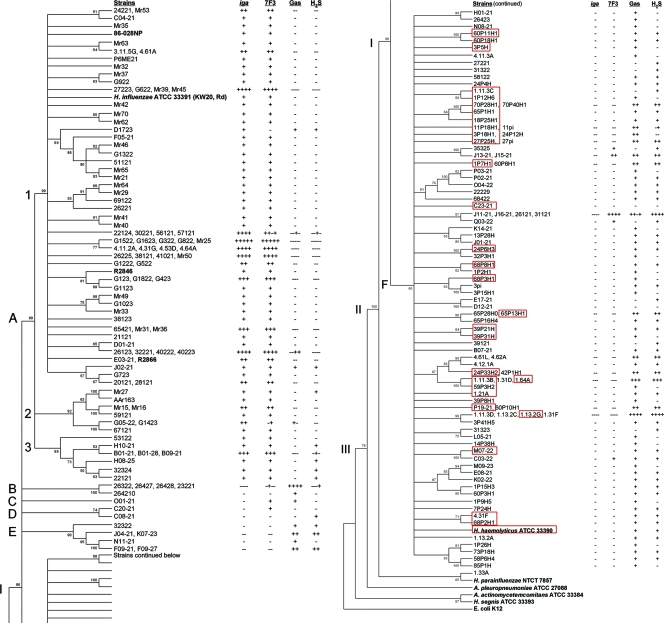
FIG. 1.
Minimum-evolution dendrogram of the Haemophilus sensu stricto cluster. The tree, rooted by E. coli strain K-12, is based on concatenated adk, pgi, recA, infB, and 16S rRNA gene sequences, with bootstrap values of ≥50% of 1,000 bootstraps indicated. Node I contains all H. influenzae and hemolytic and nonhemolytic H. haemolyticus strains within an Haemophilus sensu stricto cluster, while nodes II and III contain selected members of the family Pasteurellaceae. Strains with identical sequences are listed on the same branch. Red boxes define beta-hemolytic H. haemolyticus strains. Positive (+) and negative (−) results for the iga gene probe hybridization, P6 OMP MAb 7F3 reactivity, the production of gas during glucose metabolism, and H2S production are shown to the right of each strain. The results are also relative to the strain order on branches with multiple strains. On the basis of the stratification of iga and other results (see the text and Table 2), it is proposed that node A contains all H. influenzae strains, while nodes B to F contain all H. haemolyticus strains.