Abstract
Between 1999 and 2006, 15 cats were diagnosed with disease attributable to a novel mycobacterial species. The infections consisted of granulomatous lesions in the skin, subcutis, and ocular or periocular tissues with an indolent but progressive clinical course. Lesions typically were found in facial regions or on the distal limbs. Cats of all ages and both sexes were affected. Infections often were challenging to treat, although they could be cured using surgery in concert with combination antimicrobial therapy. Microscopically, lesions were granulomatous to pyogranulomatous and contained numerous acid-fast bacilli. Scanty cultures of the causal microorganisms occasionally could be obtained in mycobacterial broth, but subculture to solid media failed. When cultures were not available, DNA was extracted from fresh tissue, lyophilized material, and formalin-fixed, paraffin-embedded tissues from lesions. PCR amplification of the 5′ end of the 16S rRNA gene and regions within four additional loci (ITS1, hsp65, rpoB, and sodA) was performed with various efficiencies using mycobacterial primers. Nucleotide sequences were unique for each locus tested. Nucleotide sequences obtained from individual cases were identical for each locus for which the amplification was successful. Phylogenetic analysis performed using concatenated partial 16S rRNA and hsp65 gene sequences indicated that this novel mycobacterial species from Victoria is a member of the Mycobacterium simiae-related group, taxonomically related to the mycobacterium causing leproid granulomas in dogs throughout the world. Based on the clustering of cases, we refer to this novel species as Mycobacterium sp. strain Tarwin.
Feline leprosy and canine leproid granuloma syndrome (CLGS; also known as canine leprosy) are terms used to describe a range of mycobacterial infections in which cats and dogs, respectively, develop granulomas in the skin or subcutis. Lesions contain variable numbers of acid-fast bacilli (AFB), which generally do not grow using standard mycobacteriological methods, even in specialist reference laboratories.
Prior to the development of molecular techniques such as PCR, canine and feline diseases caused by fastidious mycobacteria were not well understood. This was, in part, due to the failure to culture consistently and thereby taxonomically classify the causal agents of disease using classical in vitro bacteriological methods. CLGS was first described in a boxer and a bullmastiff from Zimbabwe in 1973, with similar reports from Australia appearing soon afterwards. Primary skin lesions consist of a single or multiple, well-circumscribed nodule(s). These lesions may appear anywhere on the dog, although usually they are located on the head, typically on the dorsal ear folds. An analysis of the partial 16S rRNA gene sequence amplified directly from infected tissue (GenBank accession no. AF144747) suggests that the causal mycobacterial species is a fastidious slow grower related to Mycobacterium simiae (12).
In contrast, feline leprosy comprises a variety of clinical conditions attributable to more than one distinct mycobacterial species. Thus, erroneously attributing all cases of feline leprosy to Mycobacterium lepraemurium hindered the development of a deeper understanding of the pathogenesis of fastidious mycobacterial infections in different feline cohorts.
With the advent of molecular methods, a combined approach based on clinical observations, traditional methodologies, and sequence analysis of 16S rRNA gene PCR amplicons indicated that feline leprosy comprised two diseases in New South Wales, Australia, and New Zealand: one was associated with the rat leprosy bacillus, M. lepraemurium (GenBank accession no. AJ279017) (11), and another with a novel, fastidious, slow-growing mycobacterial species (GenBank accession no. AJ294740 to AJ294746) (13, 17). The diseases caused by these mycobacterial species differed in a number of clinical and microscopic features. Key differences related to the age and domicile of the cat, the length of AFB in lesions, the ability of bacilli to take up hematoxylin stain, the presence or absence of necrosis in lesions, and whether the pathological picture was lepromatous or tuberculoid. With one exception, culture from all Australian feline leprosy cases had been unsuccessful, despite the use of a range of appropriate media and the presence of abundant AFB in most specimens. Recent work from North America has indicated that additional, closely related mycobacterial taxa (belonging to a species provisionally called “M. visibilis”; later corrected to “M. visibile”) also cause disseminated skin disease in cats (2, 4, 8).
The Australian research included a case of unknown significance. From this case, a 2-year-old cat (named Nina) domiciled in rural Victoria, a further mycobacterial 16S rRNA sequence that did not match any database sequence was obtained (17). The lesions in this cat also were associated with numerous AFB (Table 1) (case 2 in reference 17). Interest in this case increased when two cases of mycobacterial keratitis subsequently were identified in cats residing in semirural Victoria (C. McCowan, J. Fyfe, A. O'Reilly, C. Hardman, and R. Stanley, submitted for publication). PCR amplification of the 5′ end of the 16S rRNA gene encompassing the hypervariable regions A and B, which are of particular value for the molecular differentiation of mycobacterial species (14, 23), was performed on DNA extracted from lesions from these cases. Sequence analysis of these amplicons revealed that they were identical to each other (GenBank accession no. DQ873337) and to the sequence obtained from Nina (17). Database searches indicated that this sequence represents a novel mycobacterial species belonging to the M. simiae-related group, sharing an identical short helix 18, which is characteristic of this subgroup within the slow-growing mycobacteria (27) but distinct from the 16S rRNA gene sequence determined by Hughes et al. (12) for the mycobacterium associated with CLGS (GenBank accession no. AF144747).
TABLE 1.
Clinical and demographic features of a cohort of cats with lesions likely to be attributable to Mycobacterium sp. strain Tarwina
| Case no., name of cat, and yr of diagnosis | Breed, age (yrs) | Gender | Clinical sign(s) | Pathology and biopsy findings (specimen available for DNA extraction) | Lifestyle | Sequence data obtained |
|---|---|---|---|---|---|---|
| 1, Nina, 1999 (case 2 in reference 17) | DSH, 2 | FN | Caudal right hock, then lateral digit on left forelimb | Numerous AFB, branching morphology, no necrosis, routine culture negative (lyophilized tissue) | Indoor/outdoor, occasional hunter | 16S rRNA, ITS1, hsp65 |
| 2, Sasha, 2002 (case 1 in McCowan et al., submitted) | DLH, 6 | FN | 5-mo history of raised corneal mass | Numerous AFB within macrophages, scanty growth in BACTEC 12B at 31°C, no growth on subculture (BACTEC broth culture) | Indoor/outdoor | 16S rRNA, ITS1 |
| 3, Susie, 2003 (case 2 in McCowan et al., submitted) | Burmese cross, 17 | FN | Cloudy eye that progressed to a thickened, red corneal mass | Numerous AFB, grew in BACTEC 12B at 31°C, no growth on subculture (fresh corneal tissue) | Predominantly indoor | 16S rRNA, ITS1 |
| 4, Centrals, 2005 | DLH, 3 | MN | Chin nodule and multiple nodules on head and feet | Many AFB in biopsy tissue (frozen tissue sample) | History of fighting | 16S rRNA, ITS1 |
| 5, Simba, 2005 | DSH, 4.5 | MN | Mouth and digit nodules | NA (digit nodule tissue) | NA | 16S rRNA, ITS1, hsp65, rpoB, sodA |
| 6, Jagula, 2005 | Abyssinian, 8 | FN | Firm, rapidly growing mass over frontal sinus; bone under the lesion was roughened | NA (MGIT broth culture) | NA | ITS1 |
| 7, Jazzy, 2005 | DLH, 4 | MN | Eyelid nodule, 1-cm diam | FNA showed macrophages with negatively stained bacilli (eyelid tissue) | Indoor/outdoor, history of cat fight with scratches | ITS1, hsp65 |
| 8, Runtie, 2005 | DSH, 3 | MN | First lesion on right front digit (8-mm nodule), second lesion on left forearm (20-mm nodule) | Biopsy showed multibacillary, lepromatous tissue (paraffin sections, forearm tissue) | Indoor/outdoor, hunter | 16S rRNA, ITS1, hsp65 |
| 9, Steel, 2005 | DLH, 5 | MN | Multiple nodules, head and feet | Biopsy showed multibacillary, lepromatous tissue (paraffin sections, head tissue) | Indoor/outdoor, hunter | 16S rRNA, ITS1, hsp65, rpoB |
| 10, Jack, 2004 | DSH, 3 | MN | Injury between digits on left rear foot turned into an ulcerated mass, removed surgically; 4 mo later developed a grape-sized enlargement of regional popliteal lymph node | Biopsy of interdigital lesion showed epithelial macrophages, lymphocytes, plasma cells, neutrophils plus AFB; FNA showed popliteal lymph node lymphocytes, plasma cells, and macrophages containing NSB (paraffin sections) | Indoor/outdoor, hunter | 16S rRNA, ITS1, hsp65, rpoB |
| 11, Zoe, 2003 | DSH, 10 | FN | 1-mo history of stridor/stertor and a soft-tissue mass above the upper incisors; granulomas in caudal nasal cavity | Masses of AFB in FNA of mass and nasal wash; no growth on routine culture (lyophilized tissue) | 16S rRNA, ITS1, hsp65, rpoB | |
| 12, Old Tom, 2006 | DSH, 6 | MN | Lumps on upper and lower eyelids | Large no. of AFB seen in tissue sections (paraffin sections) | 16S rRNA, ITS1, hsp65, rpoB | |
| 13, Pepper, 2006 | DSH, 13 | MN | Swelling (4 mm) lateral to nose with ipsilateral epiphora | Biopsy showed abundant epithelioid macrophages, giant cells, packed with AFB (paraffin sections) | Mainly outdoor, hunter | ITS1 |
| 14, Fang, 2004 | DSH, 2 | FN | Lump on left forelimb and left carpal area (2-cm diam), dermal plaque with ulcerated surface | FNA of lesion showed macrophages containing NSB (NA) | Indoor/outdoor, known hunter with access to rats and mice | NA |
| 15, Hoover, 2005 | DMH, 13.5 | MN | Mass on mandible just under chin (1-cm diam), painful, domed, hairless, smooth | FNA of lesion showed neutrophils and macrophages containing NSB (NA) | Mainly outdoor | NA |
Genetic loci for which nucleotide sequence information were obtained also are shown. Abbreviations: NA, not available; DSH, domestic short haired; DLH, domestic long haired; DMH, domestic medium haired; MN, male neutered; FN, female neutered; NSB, negatively stained bacilli; FNA, fine-needle aspirate.
This finding prompted a study of feline mycobacterial cases from Victoria to gain more insight into what appeared to be a potentially novel mycobacterial species as a further causative agent of feline leprosy. Clinical information for an additional 12 cases has come to our attention. These cats were from rural or semirural Victoria, while one additional cat was diagnosed in Sydney, New South Wales, Australia. Material for DNA extraction was available from 10 of these cases. The partial 16S rRNA sequence derived from the novel mycobacterial species detected in the three previous cats from Victoria was identified subsequently from seven additional patients. This paper presents molecular and/or clinical data for all 15 cases so far identified, including DNA sequence information obtained for fragments of four additional loci: internal transcribed spacer region 1 (ITS1), the β-subunit of RNA polymerase (rpoB), the gene encoding superoxide dismutase (sodA), and the gene encoding the 65-kDa heat shock protein (hsp65). Based on the multiple-gene approach described by Devulder et al. (5), we present a phylogenetic scheme for this novel species within the M. simiae-related group, including the mycobacterial species associated with CLGS. Based on the geographic location of several cases, including the initial case described by Malik et al. (17), we refer to this novel species as Mycobacterium sp. strain Tarwin.
MATERIALS AND METHODS
Case descriptions.
Mycobacterial infections of the skin, subcutis, conjunctiva, or eyelid were seen in 15 cats over an 8-year period, from 1999 to 2006 (Table 1). Domestic cross-bred cats (short to long haired) accounted for 13 of 15 cats, with the remaining two cats comprising an Abyssinian and a Burmese cross-breed.
Fourteen of the cats were domiciled in rural or semirural Victoria, the distribution of cases being illustrated in Fig. 1. Five (cases 1, 7, 10, 14, and 15) were from the South Gippsland/Tarwin region of southern Victoria; these cases had been diagnosed by a single veterinary practice, including the index case, Nina, which was reported previously. The remaining cat lived in Sydney in an area abutted by national parks. At least 10 of the cats had access to the outdoors; two were described as mainly outdoors cats, while eight were described as having an indoor/outdoor lifestyle. Six cats were known to hunt, while at least two cats were known to fight with other cats.
FIG. 1.
Geographical distribution of cases associated with Mycobacterium sp. strain Tarwin. Case numbers refer to those in Table 1. The single case from New South Wales (case 11) is not illustrated on this map of Victoria.
Of the 15 affected cats, six were spayed females and nine were castrated males. The age of affected cats ranged from 2 to 17 years, with a median age of 4.5 years. Eleven of the cats were ≤8 years of age. Lesions involved the tissues of the head (11 cases), including ocular and periocular structures (4 cases), and the distal limbs (7 cases). Three cats had involvement at multiple sites. Although limited data were retrievable from the available case notes, the disease appeared to have an indolent course, with lesions being evident from several weeks to 5 months before definitive veterinary investigations provided a diagnosis or a specific antimycobacterial therapy was initiated.
Typically, lesions consisted of nodules in the subcutis and skin. Nodules could be single or multiple (typically 8- to 20-mm diameter) (Fig. 2). In only one case was there ulceration, and in this patient the infection also spread to the regional (popliteal) lymph node. The case with a lesion originally on the nasal philtrum was unusual in that the infection subsequently spread to the posterior nasal cavity and nasopharynx. On the limbs, lesions tended to have a distal distribution, affecting the digits (three cases), antebrachium (one case), and hock (one case). Three cats had lesions on both the head and distal limbs, although the chronology of lesions was more compatible with that of simultaneous infection of multiple sites rather than dissemination via blood or lymphatic spread.
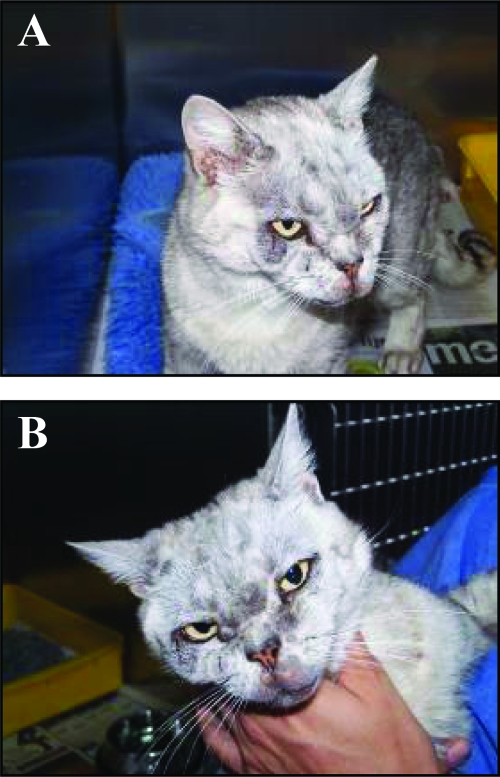
FIG. 2.
Mycobacterial granulomas due to Mycobacterium sp. strain Tarwin on the periocular skin of case 9. The lesions take the form of multiple nodules in the subcutis. Lesions are neither hot nor painful, and in contrast to M. lepraemurium lesions, ulceration was not a feature. The distribution is consistent with the introduction of bacilli on the claws of a perpetrator as a result of a cat scratch. Images A and B show different perspectives to illustrate the extent and number of the lesions.
The cases were treated by a number of different veterinarians of variable surgical expertise and using a variety of adjunctive anti-infective agents. Variable levels of owner motivation and compliance were an additional complicating issue. Furthermore, accurate follow-up of cases sometimes was impossible to obtain. The response to therapy therefore was difficult to assess accurately. The recurrence of lesions after several months often occurred at the margin after resection. Cases that were successfully and permanently cured often required two or more attempts at resection, followed up with one or more drugs with broad antimycobacterial activity, such as clarithromycin, rifampin, clofazimine, and doxycycline.
Specimens.
Table 1 shows the specimen types available for molecular analysis for each of the feline cases. Fresh, frozen, lyophilized, or formalin-fixed and paraffin-embedded tissue sections were received from 11 of the 15 cases, with scanty growth in BACTEC 12B or mycobacterium growth indicator tubes (MGIT) being provided for an additional two cases. Clinical material was not available for cases 14 and 15, so a definitive molecular diagnosis was not possible; however, based on the characteristic distribution of lesions and the geographical location of the patients, it was considered highly likely that a causative microorganism similar to that determined for the other cases was responsible. Lyophilized tissue specimens from two dogs diagnosed with CLGS were included for comparative studies.
Cytology and histopathology.
Aspirates or crush preparations from lesions were sprayed onto a glass slide, air dried, fixed in methanol, subsequently stained using a rapid Romanowsky type stain (DiffQuik; Lab Aids Australia), and examined using conventional light microscopy. Biopsy specimens were fixed in neutral buffered formalin for 12 to 24 h, embedded in paraffin, sectioned at 8 μm, and processed for conventional light microscopy. Sections were routinely stained with hematoxylin and eosin, the Ziehl-Neelsen (ZN) method, and, on occasion, with Brown and Brenn's modification of the Gram stain. The length of AFB was determined using an eyepiece graticule.
Mycobacterial culture.
Culture of mycobacteria from a fresh corneal tissue biopsy specimen received from case 3 was attempted using a variety of liquid and solid media, including MGIT (Becton Dickinson and Company, MD), Middlebrook 7H12 mycobacterial culture vials (BACTEC 12B; Becton, Dickinson, and Company), Lowenstein-Jensen medium (with and without 1% ferric ammonium citrate supplement), Brown and Buckle agar medium, chocolate agar, and buffered charcoal yeast extract medium.
DNA extraction. (i) Paraffin-embedded tissue sections.
At least six 10- to 20-μm paraffin sections in a 1.5-ml tube were dewaxed by extraction with 1 ml of histolene (Fronine Pty. Ltd.), followed by the addition of 1 ml of absolute ethanol and centrifugation at 13,000 rpm for 5 min in a microcentrifuge (Heraeus). DNA was extracted from the tissue pellet using the QIAamp DNA minikit (Qiagen Inc., Valencia, CA) by following the DNA cleanup protocol, preceded by 24 h of incubation at 56°C in digestion buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 0.5% [wt/vol] sodium dodecyl sulfate, 50 mM NaCl, 300 μg/ml proteinase K).
(ii) Fresh, frozen, and lyophilized tissue.
Tissues were diced with a scalpel and vortexed for at least a minute in a sterile 0.5-oz glass bottle containing 50 to 100 3-mm glass beads (Ajax Finechem, Seven Hills, New South Wales, Australia) and 2 ml of phosphate-buffered saline (PBS). One milliliter of the resulting cell suspension was transferred to a 1.5-ml tube and centrifuged, and DNA was extracted as described above.
PCR amplification.
Forward (F) and reverse (R) oligonucleotide primers used for PCR amplification of the various genomic DNA regions performed in this study are described in Table 2. The amplification of the ITS1 region was performed using a nested PCR protocol. Five microliters of extracted DNA was added to 20 μl of the first-round PCR mixture, with the final 25-μl volume containing PCR buffer with 1.5 mM MgCl2 (Qiagen Inc., Valencia, CA), 200 μM deoxynucleoside triphosphates (dNTPs), primers Ec16S.1390p and Mb23S.44n (at 2 μM each), and 1.25 U of Taq DNA polymerase (Qiagen). The amplification was performed with a Mastercycler gradient (Eppendorf) and the following profile: 95°C for 5 min, followed by 38 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min. The second-round PCR mixture contained 2.5 μl of the first-round PCR product in a final 50-μl volume containing PCR buffer with 1.5 mM MgCl2 (Qiagen), 200 μM dNTPs, primers SP1 and SP2 (at 2 μM each), and 2.5 U Taq DNA polymerase (Qiagen). The amplification was performed using the following profile: 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 59°C for 30 s, and 72°C for 30 s, with a final extension of 72°C for 5 min. The assay included an extraction process negative control, a PCR negative control, a positive control containing 100 fg of DNA extracted from Mycobacterium tuberculosis H37Rv, and controls for inhibition, for which tubes containing extracted DNA were spiked with 100 fg of M. tuberculosis DNA.
TABLE 2.
Oligonucleotide primers used in this study
| Gene | Primer | Directiona | Sequence (5′-3′) | Reference or source |
|---|---|---|---|---|
| ITS1 (first round) | Ec16S.1390p | F | TTG TAC ACA CCG CCC GTC A | 9 |
| Mb23S.44n | R | TCT CGA TGC CAA GGC ATC CAC C | 9 | |
| ITS1 (second round) | SP1 | F | ACC TCC TTT CTA AGG AGC ACC | 24 |
| SP2 | R | GAT GCT CGC AAC CAC TAT CCA | 24 | |
| 16S rRNA | 246 | F | AGA GTT TGA TCC TGG CTC AG | 3 |
| MR3 | R | CCT ACG AGC TCT TTA CG | This study | |
| hsp65 | Tb11 | F | ACC AAC GAT GGT GTG TCC AT | 22 |
| Tb12 | R | CTT GTC GAA CCG CAT ACC CT | 22 | |
| rpoB | MF | F | CGA CCA CTT CGG CAA CCG | 15 |
| TBB rpoB2 | R | TAC GGC GTC TCG ATG AAS CC | 5 | |
| sodA | Z205 | F | ACG TTC ACC ACA GCA AGC ACC A | 5 |
| GSOD2 | R | TCG GCC AGT TCA CGA CGT TCC A | 5 |
F, forward; R, reverse.
The amplification of a 552-bp fragment at the 5′ end of the 16S rRNA gene was performed with primer set 246 and MR3 (Table 2). Five microliters of extracted DNA was added to 45 μl of the PCR mixture, with the final 50-μl volume containing PCR buffer with 1.5 mM MgCl2 (Qiagen), Q solution (Qiagen), 200 μM dNTPs, primers 246 and MR3 (at 100 nM each), and 2.5 U Taq DNA polymerase (Qiagen). The amplification was performed using the following profile: 95°C for 2 min, followed by 40 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 5 min.
Amplifications of the hsp65, rpoB, and sodA regions were performed using the primer sets described in Table 2 and using the conditions described by Devulder et al. (5).
PCR product analysis. (i) Agarose gel electrophoresis.
PCR products were electrophoresed through 2% (wt/vol) agarose gels, and fragment sizes were estimated via comparison with a Gene Ruler 1-kb DNA ladder (MBI Fermentas, Amherst, NY) or pUC19/Msp1 marker 23 (MBI Fermentas). Fragments were purified from agarose gel slices using the High Pure PCR product purification kit (Roche) according to the manufacturer's instructions.
(ii) DNA sequence analysis.
Sequence analysis of purified PCR products was performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The primers used were the same as those described for PCR amplification (Table 2). Reactions were analyzed on a 3730S genetic analyzer (Applied Biosystems). Sequence data were edited using Bionumerics v4.0 (Applied Maths, Gent, Belgium), and database searches of GenBank were performed using the BLASTN algorithm (1).
Phylogenetic analysis.
Sequences were aligned using ClustalX (26) and were bootstrapped (1,000 replicates). Neighbor-joining phylogenetic trees were constructed using the Phylip Seqboot, DNADist/ProtDist, Neighbor, and Consense programs (http://evolution.gs.washington.edu/phylip.html). Maximum-likelihood analysis was performed using BioEdit (10), and unrooted trees were generated using TreeView (21).
Nucleotide sequence accession numbers.
The DNA sequences determined for Mycobacterium sp. strain Tarwin, isolate Simba, have been deposited in GenBank; the accession number for the partial 16S rRNA gene sequence (499 nucleotides [nt]) is EF611172, and the accession numbers for the partial ITS1 region (182 nt), partial hsp65 coding region (401 nt), partial rpoB coding region (398 nt), and partial sodA coding region (434 nt) are EF611174, EF611173, EF611175, and EF611176, respectively. The sequences determined for Mycobacterium sp. strain CLGS, isolate Metcalfe, also have been deposited in GenBank; the accession number for the partial ITS1 region (183 nt) is EF611177, and the accession number for the partial hsp65 coding region (401 nt) is EF611178.
RESULTS
Cytology and histopathology.
All cases were provisionally diagnosed as mycobacterial infections on the basis of detecting negatively staining bacilli in cytological preparations stained with DiffQuik (5 cases), AFB in cytological smears (1 case), or histological sections (10 cases) obtained from lesions and subjected to ZN staining. For all cases for which a pathological description could be retrieved, multibacillary, likely lepromatous, disease was recorded in the patient notes, with samples from the cases exhibiting sheets of plump macrophages with only sparse lymphocytes and neutrophils. Negatively stained bacilli and AFB were plentiful, were estimated to be between 2 and 3.5 μm in length, failed to take up hematoxylin in hematoxylin-and-eosin-stained sections, and commonly were arranged in haphazard clusters within macrophages. Beading of AFB was rare. In one case, AFB were described as having a branching morphology. Caseous necrosis was not observed in any of the lesions.
Mycobacterial culture.
Mycobacteria from fresh corneal tissue received from case 3 were cultured in the BACTEC 12B system at 31°C (Fig. 3) . After 1 week, abundant AFB were seen following ZN staining of a smear prepared from the broth culture; however, attempts to further subculture the bacilli using a range of different media and conditions were unsuccessful. Incubation temperatures of 31, 36, and 43°C, as well as culturing under microaerophilic conditions, failed to yield visible growth on any of the solid media following 12 weeks of incubation. Scanty growth in BACTEC 12B likewise was obtained for case 2, and scanty growth in an MGIT was supplied for case 6 by the referring pathology laboratory. Further subculture again failed to yield mycobacterial growth.
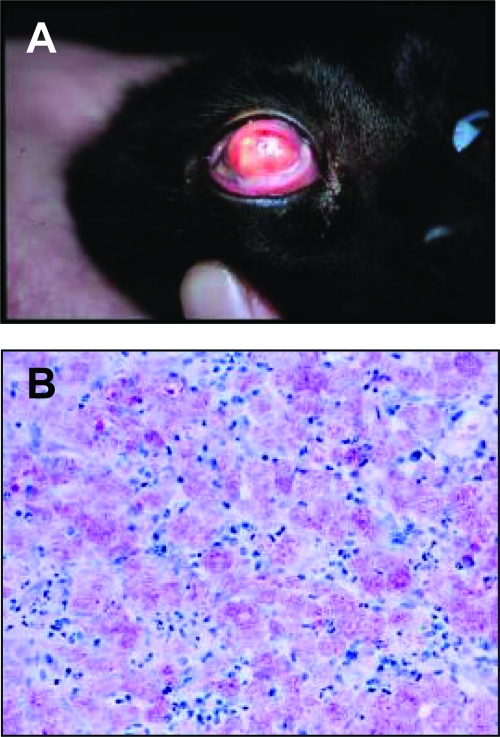
FIG. 3.
Mycobacterial keratitis and conjunctivitis due to Mycobacterium sp. strain Tarwin. The gross appearance of the lesions are illustrated (A), while the photomicrograph stained with ZN illustrates granulomatous inflammation with epithelioid macrophages containing enormous numbers of AFB (B).
Molecular analysis.
For all 13 cases for which biopsy material or cultures were available for DNA extraction (Table 1), a 224-bp product was amplified in the second round of the PCR targeting the ITS1 region of the mycobacterial genome. The nucleotide sequences of the 182-bp fragment flanked by the primers SP1 and SP2 were identical for all of the products, and a BLASTN search of the GenBank nr databases revealed the highest nucleotide identity (95%) with the ITS1 region of “Mycobacterium tilburgii” (accession no. AJ580827) (16) and 93 to 94% similarity to other members of the M. simiae-related group. A 225-bp PCR product likewise was amplified from DNA extracts from two dogs diagnosed with CLGS. This sequence also shared 95% identity with the ITS1 sequence from M. tilburgii and shared 94.5% identity with the ITS1 sequence obtained from the isolates of Mycobacterium sp. strain Tarwin.
In addition to the three cases for which the 5′ end of the 16S rRNA gene previously had been analyzed, a PCR product was amplified from a further seven cases. The sequences in all 10 instances were identical to that of GenBank accession no. DQ873337 and shared the closest identity (99.2%) with the 16S rRNA gene sequence from “Mycobacterium sherrisii” (GenBank accession no. AY353699) (25) and Mycobacterium sp. strain IP20010510 (GenBank accession no. AY163334), an environmental isolate from water. Figure 4 shows an alignment of sequences within helix 18 between several of the Mycobacterium species previously known to cause feline leprosy syndrome or disseminated disease in cats (M. lepraemurium; Mycobacterium sp. strain cat; M. visibile isolates Idaho, Oregon, and Saskatchewan; Mycobacterium sp. strain Tarwin; 15 other M. simiae-related mycobacteria [including the CLGS-associated mycobacterium]; and M. tuberculosis). This alignment shows that all of these feline Mycobacterium species can be distinguished from each other on the basis of the nucleotide sequence within helix 18. Unlike the others, Mycobacterium sp. strain Tarwin has the characteristic insertion-free version of this region shared with other members of the M. simiae-related mycobacterial species. However, relative to the other M. simiae-related mycobacteria, the novel species has a C-T substitution at nt 467 (the numbering is based on that of the M. tuberculosis 16S rRNA gene sequence).
FIG. 4.
Nucleotide sequence alignment within helix 18 of the V3 region of the 16S rRNA gene that compares the sequence determined for M. tuberculosis to the sequences determined for the Mycobacterium species associated with feline leprosy and disseminated disease in cats. Note that Mycobacterium sp. strain Tarwin (M. sp. Tarwin) has the insertion-free version, relative to the other slow-growing mycobacteria, characteristic of the M. simiae-related Mycobacterium species. Note also the C-T substitution (in boldface) at nt 467, relative to the other M. simiae-related species. Superscript letters a to f indicate the following GenBank accession numbers: a, BX842576; b, AJ279017; c, AJ294740 to AJ294746; d, AY061986; e, AY061985; f, AY061984. M. simiae-related refers to the following species: M. simiae, M. genavense, M. lentiflavum, M. triplex, M. interjectum, M. intermedium, M. heidelbergense, M. parascrofulaceum, M. kubicae, M. palustre, M. montefiorense, M. florentinum, M. sherrisii, Mycobacterium sp. strain IP20010510, and Mycobacterium sp. strain CLGS (M. sp. CLGS).
A 441-bp PCR product within the hsp65 gene was amplified and sequenced from DNA extracted from 8 of the 13 cases. Again, all sequences were identical. A BLASTN search of the nr databases indicated that the 401-nt region flanked by the primers shared 97% identity with sequences obtained for M. simiae strains ATCC 25275 and CIP 104531 (GenBank accession no. AF434730 and AF547875, respectively). A 441-bp product within hsp65 likewise was amplified and sequenced from DNA extracted from the two CLGS cases. These sequences were identical to each other but shared only 94.3% nucleotide identity with the sequence determined for Mycobacterium sp. strain Tarwin.
Amplification of a 436-bp fragment within the rpoB gene was successful in five cases (all sequences were identical), with a database search using the 398-nt region flanked by the primers revealing 95% identity with the rpoB sequence from M. simiae strain CIP 104531 (GenBank accession no. AY44963). However, only for the DNA extract from case 5 was a product amplified from within the sodA gene, using primers Z205 and GSOD2. This sequence shared 92% identity with the comparable region of the sodA gene from M. simiae CIP 104531 (GenBank accession no. AY544864). The amplification of the rpoB and sodA regions unfortunately was unsuccessful for the DNA extracts from the two CLGS cases using the primers and amplification conditions described in Materials and Methods. Consequently, the rpoB and sodA sequences were not included in the phylogenetic analysis so that the Mycobacterium sp. strain CLGS could be included.
Phylogenetic analysis.
Although there are many partial 16S rRNA gene sequences deposited in GenBank that had high nucleotide identity to the sequences determined for Mycobacterium sp. strain Tarwin and Mycobacterium sp. strain CLGS, there are fewer hsp65 sequences available for comparison. For the purposes of phylogenetic analysis, we decided to include only the M. simiae-related species for which there were paired 16S rRNA and hsp65 sequences from the same isolate (Table 3). The partial 16S rRNA gene sequences corresponding to nt 1 to 499 of the sequence with GenBank accession no. AF547966 and hsp65 sequences corresponding to nt 14 to 414 of GenBank accession no. AF547875, determined for the M. simiae isolate CIP104531, were selected for each species. An analysis of the concatenated 16S and hsp65 sequences showed that all major clades received strong bootstrap support, with the strongest support observed for the clade containing M. simiae, Mycobacterium sp. strain Tarwin, Mycobacterium sp. strain CLGS, and M. sherrisii (Fig. 5). This suggests a common evolutionary origin for these novel species, distinguishing them from the other members of the M. simiae-related mycobacteria. Phylogenetic trees with similar topology but reduced bootstrap support were generated using the 16S rRNA gene and hsp65 sequences independently (data not shown).
TABLE 3.
Mycobacterial species, strains, and accession numbers of sequences used in the phylogenetic analysis
| Mycobacterial species and strain no. | Accession no. for:
|
|
|---|---|---|
| 16S rRNA gene | hsp65 | |
| Mycobacterium sp. strain Tarwin, isolate Simba | EF611172 | EF611173 |
| Mycobacterium sp. strain CLGS, isolate Metcalfe | AF144747a | EF611178 |
| M. florentinum DSM 44852 | DQ350154 | DQ350162 |
| M. genavense DSM 44424 | AF547928 | AF547837 |
| M. heidelbergense CIP 105424 | AF547935 | AF547844 |
| M. interjectum DSM 44064 | AF547937 | AF547846 |
| M. kubicae CIP 106428 | AF547941 | AF547850 |
| M. lentiflavum CIP 105465 | AF547942 | AF547851 |
| M. palustre DSM 44572 | AY943210 | AY943200 |
| M. parascrofulaceum CIP 108112 | AY943211 | AY943201 |
| M. sherrisii ATCC BAA-832 | AY353699 | AY365190 |
| M. simiae CIP 104531 | AF547966 | AF547875 |
| M. triplex CIP 106108 | AF547973 | AF547882 |
FIG. 5.
Unrooted phylogenetic tree of the M. simiae-related mycobacterial species, including Mycobacterium sp. strain Tarwin and Mycobacterium sp. strain CLGS, computed from the concatenated 16S rRNA gene and hsp65 sequences by the neighbor-joining method.
DISCUSSION
This paper provides the first molecular characterization of a novel etiological agent of feline leprosy, one caused by a fastidious mycobacterial species (Mycobacterium sp. strain Tarwin) that, like the microorganism that causes CLGS in dogs, is a member of the M. simiae-related group. A phylogenetic analysis based on the concatenated partial 16S rRNA and hsp65 gene sequences supports the notion that these agents represent separate mycobacterial species with a common evolutionary origin. Sequence comparison of a third locus, the ITS1 region, further supports a distinction between Mycobacterium sp. strain Tarwin and Mycobacterium sp. strain CLGS, as the sequences share only 94.5% identity. The nested PCR targeting the ITS1 region successfully amplified products for all of the cases for which appropriate material was available for DNA extraction, whereas the single-round protocols targeting the other loci were less successful, probably due to reduced sensitivity.
Feline leprosy is a heterogeneous syndrome comprising a number of different diseases. To date, etiological agents include M. lepraemurium, the novel Victorian species described here (Mycobacterium sp. strain Tarwin), the unnamed novel species described in New South Wales, Australia, and New Zealand, and M. visibile, which was isolated from cats in North America.
The distribution of lesions, on the facial regions and distal limbs, in cases attributable to this Victorian species implicates a traumatic pathogenesis (6, 18, 19). The development of disease likely involves an interplay between the mycobacterial inoculum and the immune response of the host. A large inoculum, deep introduction into the tissues, and the concurrent introduction of foreign vegetable matter all would favor the establishment of infection, especially in a host with an immunological makeup inherently less adept at dealing with mycobacteria capable of intracellular survival. Lesions in an affected cat appear to remain localized at first, with subsequent extension to nearby adjacent tissues and lymphatic spread. In some cases, lesions appeared sequentially at disparate anatomic locations (e.g., digit, hock, and head), suggesting initial deposition of bacilli at multiple sites, but with some lesions developing more quickly than others. A similar phenomenon occurs in CLGS, except it is thought that biting flies act as mechanical vectors and inoculate bacilli from an environmental niche into disparate locations (12).
The clinical course in most cases was indolent but progressive. There were no instances of spontaneous remission, in contrast to CLGS, for which self cure is common. Ulceration was observed in only one instance. Histologically, the disease was typically multibacillary and lepromatous. Such a picture is generally suggestive of host immune deficiency. Studies of the feline immunodeficiency virus status of affected cats and their CD4 lymphocyte subset count would be informative in relation to this issue. Another possibility is that cats with a defective immunological makeup, possibly related to major histocompatibility complex polymorphisms, are at increased risk for developing mycobacterial disease.
Clinically, the disease caused by Mycobacterium sp. strain Tarwin had many features in common with M. lepraemurium infections, such as a stereotyped distribution of lesions. However, the indolent clinical course and multibacillary, lepromatous pathology are more typical of disease referable to the novel New South Wales and New Zealand Mycobacterium species (GenBank accession no. AJ294740 to AJ294746) (17), although lesions were not as numerous and cats of a wide age range were affected, in contrast to the older cohort of cats affected by the latter species.
One of the most remarkable features of this story was the geographical proximity of patients, with all but one case occurring within a limited part of rural/semirural Victoria (Fig. 1). Indeed, five cases were diagnosed by a single veterinary group. The geographical proximity of cases stands in contrast to the closely related organism that causes CLGS, which has a ubiquitous worldwide distribution based on an identical partial 16S rRNA sequence for isolates from North America, Hawaii, Australia, and Brazil (7).
Cats are an excellent sentinel species for environmentally acquired infections, as they have a finite geographical range (4, 20). It is therefore highly likely that Mycobacterium sp. strain Tarwin has an environmental niche that is much more common or accessible to cats in this location than elsewhere. It is our impression that the disease has become more common over the study period, with 8 of the 15 cats having first developed clinical signs in 2005 or 2006. This may be attributable to the amplification of mycobacteria in their environmental niche as a result of unknown ambient conditions (temperature, humidity, rainfall, etc.).
A further challenge is the determination of conditions that will permit these fastidious mycobacteria to grow on synthetic media. It is possible that they are capable of only intracellular growth and are able to be cultured in association with another microorganism, such as an amoeba.
The evolving picture of mycobacterial disease in companion animals shows the critical importance of molecular techniques. The finding of a single discordant 16S rRNA sequence in our early studies initially was a cause of great concern, as at that time we were not prepared to believe that three different mycobacterial species could be involved in feline leprosy. Indeed, we suspected a methodological error. The subsequent discovery of 14 additional cases, all but one from consistent geography, provided compelling evidence that a third species of Mycobacterium was involved, and the somewhat unique distribution of lesions further supported this contention. Considering the one (or two) new species reported by Appleyard and Clarke (2) and Foley et al. (8), it is clear that cats are susceptible to a wide range of fastidious mycobacterial species that can produce lesions that are virtually impossible to distinguish without molecular verification. This poses a significant challenge for the clinician, as in the absence of culture and in vitro susceptibility testing it will be necessary to empirically determine the most effective therapy for each species.
Acknowledgments
We thank the numerous veterinarians who contributed clinical material and their case notes. In particular, we thank the Tarwin veterinary group, and Caroline Bisset in particular, for their help with this story. Patricia Martin assisted in some of the cytological observations, Denise Wigney helped maintain our archive of mycobacterial specimens, and Paul Canfield assisted with pathology. Thanks also go to the late Daria Love, who instigated Australian studies into feline leprosy in the 1970s.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleyard, G. D., and E. G. Clarke. 2002. Histologic and genotypic characterization of a novel Mycobacterium species found in three cats. J. Clin. Microbiol. 402425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Bottger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 281751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, J. L., J. A. Sibley, S. Myers, E. G. Clark, and G. D. Appleyard. 2006. Histological and genotypical characterization of feline cutaneous mycobacteriosis: a retrospective study of formalin-fixed paraffin-embedded tissues. Vet. Dermatol. 17155-162. [DOI] [PubMed] [Google Scholar]
- 5.Devulder, G., M. Perouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55293-302. [DOI] [PubMed] [Google Scholar]
- 6.Deykin, A. R., D. I. Wigney, J. S. Smith, and B. D. Young. 1996. Corneal granuloma caused by Mycobacterium intracellulare in a cat. Aust. Vet. Practit. 2623-26. [Google Scholar]
- 7.Foley, J. E., D. Borjesson, T. L. Gross, C. Rand, M. Needham, and A. Poland. 2002. Clinical, microscopic, and molecular aspects of canine leproid granuloma in the United States. Vet. Pathol. 39234-239. [DOI] [PubMed] [Google Scholar]
- 8.Foley, J. E., T. L. Gross, N. Drazenovich, F. Ramiro-Ibanez, and E. Anacleto. 2005. Clinical, pathological and molecular characterisation of feline leprosy syndrome in the western USA, p. 238-246. In A. Foster, K. Kwochka, G. Bertola, and A. Hillier (ed.), Advances in veterinary dermatology, vol. 5. Blackwell Publishing Ltd., Oxford, United Kingdom. [Google Scholar]
- 9.Frothingham, R., and K. H. Wilson. 1993. Sequence-based differentiation of strains in the Mycobacterium avium complex. J. Bacteriol. 1752818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 4195-98. [Google Scholar]
- 11.Hughes, M. S., N. W. Ball, L.-A. Beck, G. W. de Lisle, R. A. Skuce, and S. D. Neill. 1997. Determination of the etiology of presumptive feline leprosy by 16S rRNA gene analysis. J. Clin. Microbiol. 352464-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, M. S., J. G. James, N. Ball, M. Scally, R. Malik, D. Wigney, P. Martin, S. Chen, D. Mitchell, and D. N. Love. 2000. Identification by 16S rRNA gene analyses of a potential novel mycobacterial species as an etiological agent of canine leproid granuloma syndrome. J. Clin. Microbiol. 38953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, M. S., G. James, M. J. Taylor, J. McCarroll, S. D. Neill, S. C. A. Chen, D. H. Mitchell, D. N. Love, and R. Malik. 2004. PCR studies of feline leprosy cases. J. Feline Med. Surg. 6235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kempsell, K. E., Y. Ji, G. Estrada, M. J. Colston, and R. A. Cox. 1992. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J. Gen. Microbiol. 1381717-1727. [DOI] [PubMed] [Google Scholar]
- 15.Kim, B. J., S. H. Lee, M. A. Lyu, S. J. Kim, G. H. Bai, G. T. Chae, C. Y. Cha, and Y. H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 371714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolditz, M., M. Halank, P. Spornraft-Ragaller, H. Schmidt, and G. Hoffken. 2005. Localized pulmonary infection associated with Mycobacterium tilburgii in an HIV-infected patient. Infection 33278-281. [DOI] [PubMed] [Google Scholar]
- 17.Malik, R., M. S. Hughes, G. James, P. Martin, D. I. Wigney, P. J. Canfield, S. C. A. Chen, D. H. Mitchell, and D. N. Love. 2002. Feline leprosy: two different clinical syndromes. J. Feline Med. Surg. 443-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik, R., J. Norris, J. White, and B. Jantulik. 2006. Wound cat. J. Feline Med. Surg. 8135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik, R., L. Vogelnest, C. R. O'Brien, J. White, C. Hawke, D. I. Wigney, P. Martin, and J. M. Norris. 2004. Infections and some other conditions affecting the skin and subcutis of the naso-ocular region of cats-clinical experience 1987-2003. J. Feline Med. Surg. 6383-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien, C. R., M. B. Krockenberger, D. I. Wigney, P. Martin, and R. Malik. 2005. Retrospective study of feline and canine cryptococcosis in Australia from 1981 to 2001: 195 cases. Med. Mycol. 42449-460. [DOI] [PubMed] [Google Scholar]
- 21.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 22.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogall, T., T. Flohr, and E. C. Böttger. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 1361915-1920. [DOI] [PubMed] [Google Scholar]
- 24.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 381094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selvarangan, R., W. K. Wu, T. T. Nguyen, L. D. Carlson, C. K. Wallis, S. K. Stiglich, Y. C. Chen, K. C. Jost, Jr., J. L. Prentice, R. J. Wallace, Jr., S. L. Barrett, B. T. Cookson, and M. B. Coyle. 2004. Characterization of a novel group of mycobacteria and proposal of Mycobacterium sherrisii sp. nov. J. Clin. Microbiol. 4252-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortoli, E., C. Piersimoni, P. Kirschner, A. Bartoloni, C. Burrini, C. Lacchini, A. Mantella, G. Muzzi, C. Passerini Tosi, V. Penati, C. Scarparo, M. Tullia Simonetti, and E. C. Bottger. 1997. Characterization of mycobacterial isolates phylogenetically related to, but different from, Mycobacterium simiae. J. Clin. Microbiol. 35697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]