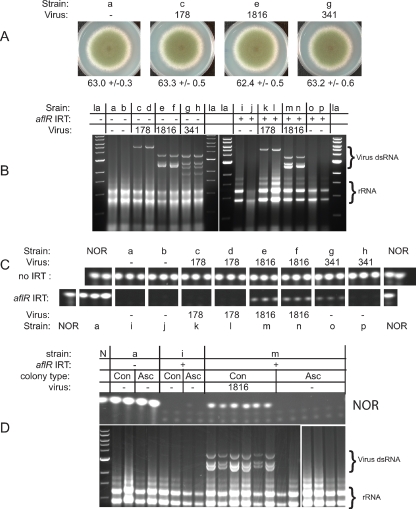
FIG. 2.
Virus 1816 suppresses RNA silencing. Strains are labeled with a lowercase letter, which can be used with the list in Table 1 to obtain the full genotype of each strain. Pertinent genotype information is provided in each panel. Radial growth and NOR production were determined for non-IRT and aflR (IRT) carrying, mycovirus-infected strains of A. nidulans. (A) None of the three mycoviruses affected radial growth or overall morphology of A. nidulans strains lacking an aflR IRT. Radial growth averages (in mm) with standard deviation values are listed (n = 5 to 6). (B) A single replica plate from the assay for each virus-strain combination was checked for mycovirus by gel electrophoresis. For virus 1816, the two shortest bands sometimes migrate as one band. Virus 341 was lost from strains o and p sometime during or before the experiment. la, DNA ladder. (C) NOR levels were determined in three replica plates for each strain-virus combination. The top row includes data from non-IRT strains, and the bottom row includes data from the corresponding aflR IRT strains. NOR levels were higher in virus 1816-infected strains (m and n) relative to control strains (i and j). The NOR level was also higher in strain o relative to the control strains (i and j), but this phenotype was unstable (see the text). (D) Single-conidium-derived (Con) and single-ascospore-derived (Asc) colonies were obtained from strains a, i, and m and analyzed for mycovirus presence and NOR production. Each lane represents an independent colony. Ascospore passage eliminated virus 1816 from strain m and restored RNA silencing-based suppression of NOR production. N, NOR standard or DNA ladder.