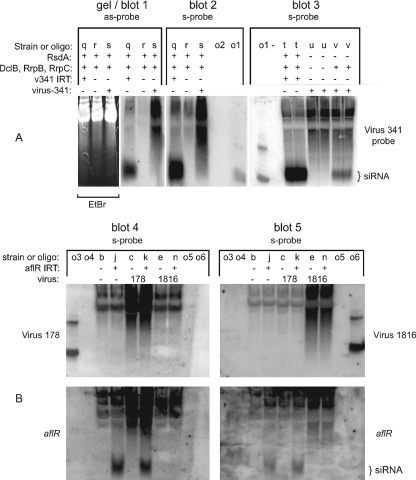
FIG. 3.
Mycovirus-derived siRNA is present in a virus 341-infected Argonaute mutant. Strains are labeled with a lowercase letter, which can be used with Table 1 to obtain the full genotype of each strain. In addition, pertinent genotype information and virus infection status are indicated above each lane with plus (+) and minus (−) symbols. (A) Low-molecular weight RNA from various A. nidulans strains was hybridized to a probe made of virus 341 sequences. Either antisense (as-probe)- or sense (s-probe)-specific riboprobes were used. For blot 1, the ethidium bromide-stained gel is shown to demonstrate relative RNA levels. siRNA is only detected in the v341 IRT strain (blots 1 to 3, strain q) and a virus 341-infected Argonaute mutant (blot 3, strain v). (B) Low-molecular-weight RNA from various A. nidulans strains was hybridized to a probe consisting of virus 178 (top left) and virus 1816 (top right) sequences, followed by a probe consisting of aflR sequences (bottom). Sense-specific riboprobes were used for each analysis. Mycovirus-derived siRNA and aflR siRNA were not detected for virus 1816, indicating that virus 1816 interferes with IRT-derived siRNA levels. Mycovirus-derived siRNA was also not detected for virus 178, but aflR siRNA was not affected by this mycovirus. For blots 2, 3, 4, and 5, 50 to 100 pmol of oligonucleotides were loaded as controls (see Fig. 1). In blots 3, 4, and 5, two bands can be seen in some oligonucleotide control lanes. It appears that the higher band is due to impurities in the oligonucleotide preparation, since it is present even when only one oligonucleotide is loaded as a control (blot 3). This also suggests that only one of the two oligonucleotides is recognized by the riboprobe in blots 4 and 5. This may be an artifact related to a decreased specific activity of the riboprobe at increasing distances from the transcription start site, thus causing the oligonucleotide closest to the transcription start site to be the only band detected by the riboprobe.