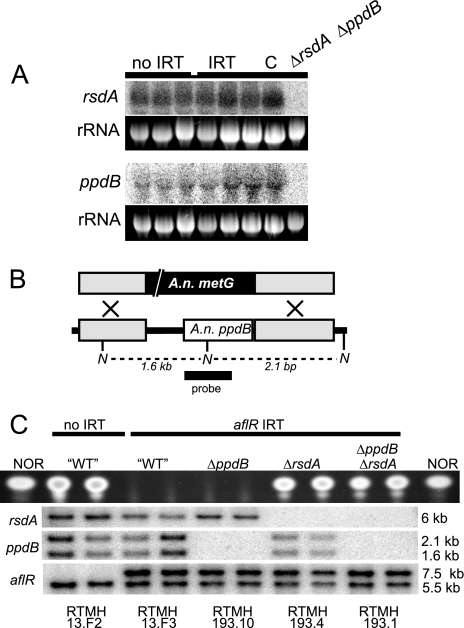
FIG. 4.
The truncated A. nidulans ppdB locus is transcribed but not required for IRT-RNA silencing. (A) Northern blotting identifies ppdB and rsdA transcripts in total RNA from 48-h cultures. The rsdA transcript migrates slightly above the 26S rRNA band, while the ppdB transcript migrates slightly below the 18S rRNA band, as expected by its truncated nature. Strains: no IRT, RTMH13.B3; IRT, RTMH 13.B1; C, RTMH192.3; ΔrsdA ΔppdB, RTMH193.1. (B) Schematic representation of ppdB replacement with A. nidulans metG. Gray boxes represent the ppdB-flanking sequences used in the deletion vector. The white box represents the predicted ppdB ORF. N, NcoI sites. (C) TLC analysis of NOR production and Southern blotting (NcoI digest for ppdB; HindIII digest for rsdA and aflR) results for control strains and recombinants from a cross between a ΔppdB transformant (TBRG3.6) and an ΔrsdA transformant (TEAB65.C1). In the Southern blots, the absence of bands for ppdB and rsdA (30) is indicative of the deletion genotypes (see probe templates in panel B and also the supplemental material), and the presence of two bands for aflR is indicative of the aflR IRT genotype. Note that RNA silencing of aflR is dependent on rsdA but not on ppdB. Strain names are listed below each lane.