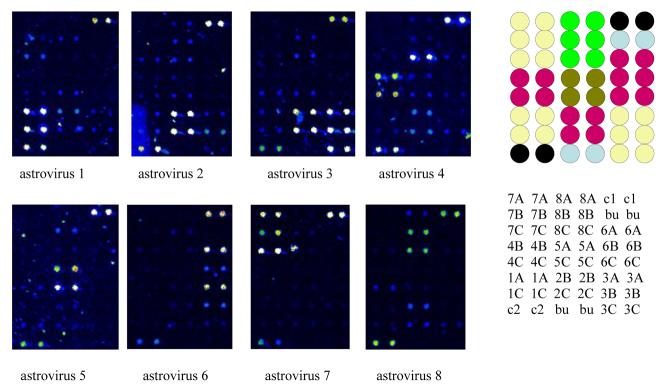
Fig. 3.
Oligonucleotide microarray for distinguishing the eight different types of human astrovirus. RT-PCR was performed using a single pair of primers at equimolar concentrations. The antisense primer was labeled with Cy3. The RT-PCR products were enzymatically digested to remove the unlabeled (and unprotected) sense strands, and the remaining labeled antisense targets were column purified and applied to the microarray consisting of predominantly 17mer positive sense probes. Duplicate dots in the upper right and lower left of each array are two conserved sequences in common to all the astroviruses. Type specific probes are clustered as two to three pairs of duplicate dots on the array. The pseudocolor scale used to indicate signal intensity is as follows: black < blue < green < yellow < orange < red < white. Locations of probes on the microarray are provided to the right of the scans. A = site 3. B = site 4. C = site 5. c1 = common sequence, site 1. c2 = common sequence, site 2. bu = buffer control.