Figure 5. 3C assay for intramolecular looping.
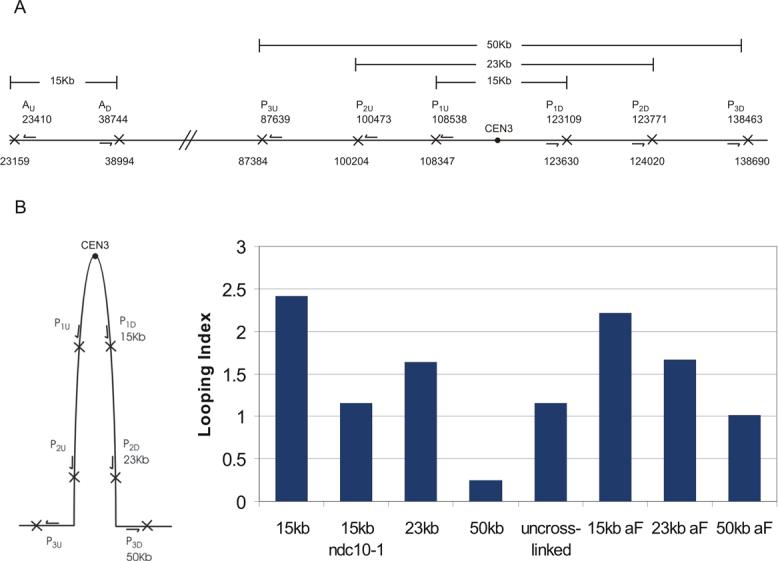
A. The schematic shows the position of oligonucleotide primers on chromosome III (arrows) relative to the centromere (filled circle). Each pair of oligonucleotides (P1u,P1d and Au,Ad; pericentric vs. arm chromatin) extend away from each other on the linear chromosome. XbaI sites are indicated by (X) downstream each oligonucleotide primer. In the linear chromosomal configuration, these oligonucleotides will not prime DNA synthesis following the 3C assay. If there is intramolecular looping (as diagrammed in B, left), the P1u,P1d oligonucleotides will prime DNA synthesis. The products from PCR reactions following cross-linking, restriction digestion and ligation were quantified as described in Material and Methods. B. (left) Schematic representation of the extent of the intramolecular loop. (right) Looping index for each experimental sample. The looping index accounts for differential efficiency of PCR reactions with primer set P versus A at an equivalent ratio of input template (see complete description in Suppl. Fig. 3). A looping index of 1 indicates equal concentration of input template for pericentric and arm products respectively. Experimental samples (WT-wild-type; alpha factor, ndc10−1, and uncrosslinked) were prepared as described in Materials and Methods.
