Abstract
Protein sequences are the most important source of evolutionary and functional information for new proteins. In order to facilitate the computationally intensive tasks of sequence analysis, the Similarity Matrix of Proteins (SIMAP) database aims to provide a comprehensive and up-to-date dataset of the pre-calculated sequence similarity matrix and sequence-based features like InterPro domains for all proteins contained in the major public sequence databases. As of September 2007, SIMAP covers ∼17 million proteins and more than 6 million non-redundant sequences and provides a complete annotation based on InterPro 16. Novel features of SIMAP include a new, portlet-based web portal providing multiple, structured views on retrieved proteins and integration of protein clusters and a unique search method for similar domain architectures. Access to SIMAP is freely provided for academic use through the web portal for individuals at http://mips.gsf.de/simap/and through Web Services for programmatic access at http://mips.gsf.de/webservices/services/SimapService2.0?wsdl.
INTRODUCTION
The number of proteins stored in public databases is rapidly growing and the sequences of amino acids are, at the moment, the most important source of evolutionary and functional information for new proteins. Therefore, the calculations of similarities and features based on protein sequences are by far the most frequently used bioinformatics applications and consume huge amounts of CPU cycles worldwide.
Database searches of individual sequences that are already included in sequence databases and the generation of sequence similarity networks by all-against-all comparisons, e.g. for clustering of proteins or prediction of orthologous groups, can be drastically accelerated and cheapened by the pre-calculation of sequence similarities and features. Redundant calculations are hence replaced by retrieval of data from a database. In order for such a database to be useful and applicable to a wide range of bioinformatics problems, it should cover the known protein space comprehensively and be frequently updated.
The database ‘Similarity Matrix of Proteins’ (SIMAP) aims to provide a comprehensive and up-to-date dataset of pre-calculated sequence similarities and features for all proteins contained in the major public sequence databases, including Uniprot/Swissprot, Uniprot/TrEMBL (1), PDB (2), GenBank (3) and RefSeq (4). Due to its high coverage and frequent update cycles, SIMAP has developed into the largest and thus unique resource of pre-calculated sequence analysis so far.
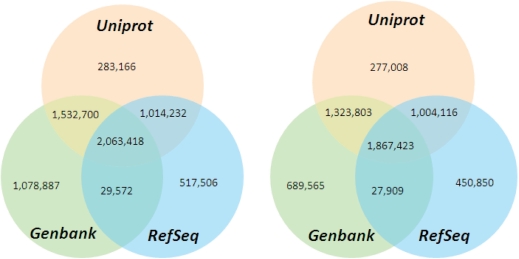
The core of SIMAP consists of a database system that consistently stores all proteins imported from heterogeneous data sources and provides efficient and fully automated update functionality (5). The amino acid sequences are kept non-redundantly, resulting in a current number of ∼17 million proteins and >6 million sequences in SIMAP (see Figure 1 for a comparison of the proteins and sequences covered by the three most important public sequence databases). The basic protein data are supplemented by the taxonomic assignments, if available, from the source databases. Other information that is important for downstream analysis, e.g. chromosomal location or functional annotation, is available from the tightly interconnected PEDANT genome database system (6).
Figure 1.
Numbers of the proteins (left) and non-redundant sequences (right) covered by the three most important public sequence databases: Uniprot, RefSeq and GenBank as of September 2007.
For all non-redundant sequences in SIMAP, a matrix of all-against-all sequence similarities [calculated by a sensitive two-step algorithm based on FASTA and Smith–Waterman (5)] is maintained by our system. In contrast to other databases storing pre-calculated similarities (like NCBI BLink), the similarity calculation is thresholded only by a static and sensitive raw score cutoff and not by a maximal number of hits per sequence. Therefore, the structure of the graph formed by the sequence similarities is not altered by the representation of particular protein families in sequence databases and is thus well suited for downstream analysis like clustering or the analysis of its network structure.
To facilitate the individual analysis of protein families, the graph formed by pairwise sequence alignments has to be complemented by position-specific scoring of similarities in order to focus on functionally or structurally important residues. SIMAP therefore provides pre-calculated predictions of protein domains for all member databases of InterPro (7) and of additional features like transmembrane helices (8), signal peptides (9) or localization predictions (10) for the complete set of sequences.
The computational space of calculating sequence similarities and features is minimized by the non-redundant representation of both sequences and feature models. This allows for a strictly incremental updating procedure, not only with respect to the sequences but also for the feature space. Thus, when upgrading all SIMAP features to a new InterPro release, only a usually small number of changed and new domain models have to be calculated. Most of the calculations are performed by the public resource computing project BOINCSIMAP (11).
All data in SIMAP are freely available for academic use through the web portal and Web Services. The smaller parts of the data, i.e. protein and sequence information and the sequence features, can be downloaded as flat files. The similarity data are not suited for direct download due to its huge size of currently more than 1 TB and can therefore only be accessed through the SIMAP Web Services. For projects that want to make use of SIMAP data for a large set of proteins, dumps are provided individually upon request, including a regular update service.
NEW FEATURES AND IMPROVEMENTS IN SIMAP
User-friendly access through integrative web portal
To retrieve proteins, features and homologs from SIMAP, a new and improved web portal provides a user-friendly and powerful toolbox. During the implementation of this portal, the integration of information from heterogeneous databases into the different views, e.g. proteins and homologs from SIMAP and functional annotation from PEDANT, has been a major handicap. Therefore, the new SIMAP web portal is based on an enterprise portal server that is capable of aggregating individual content by reusable portlets, thereby providing context-specific views.
The entry point into SIMAP through the web portal is to search proteins by user-defined text terms and sequences. If a query sequence cannot be found in SIMAP, the closely related sequences are searched by a rapid ‘SeqFinder’ algorithm based on a suffix array representation of SIMAP. In order to find related sequences in the SIMAP database, the query sequence is translated into a reduced alphabet of 10 groups of amino acids having positive substitution scores in the BLOSUM50 matrix (12). The transformed query sequence is fragmented into overlapping short substrings. Each substring is searched for exact matches in the suffix array representation of SIMAP, which also has been transformed into the reduced alphabet of amino acids (13). All matching sequences are classified by their relation to the complete query sequence as ‘equal’, ‘containing’, ‘contained’ and ‘similar’ sequences. The search space can be reduced easily by selection of databases and taxa. The classical list view of the results is complemented by a taxonomic view, which allows the user to explore the proteins found in a tree-like structure based on by the NCBI taxonomy (14).
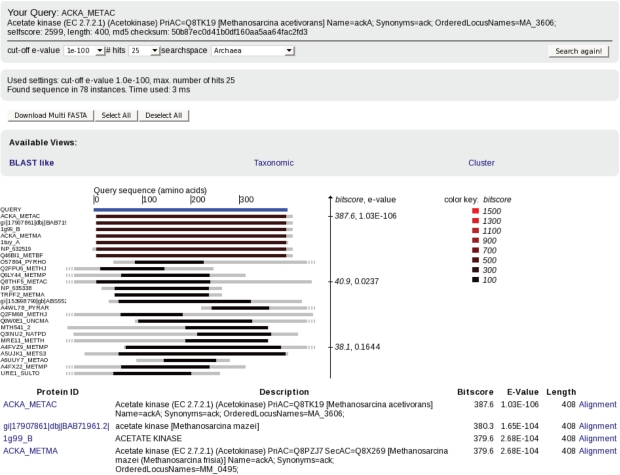
For every protein in SIMAP, its pre-calculated features and the list of homologs can be retrieved immediately from the database. To explore homologous proteins by multiple criteria, the classical result list including a graphical representation of the alignments and grouping of proteins that share the same sequences (Figure 2) is complemented by alternative views that structure the homologs by taxonomy or assignment to sequence clusters (see below).
Figure 2.
To explore the homologs of a user-defined query protein, the classical result list including a graphical representation of the alignments is shown per default. Additional views allow structuring the homologs by taxonomy or assignment to sequence clusters.
Structuring the sequence space by clustering of protein families
In order to structure the sequence space of known proteins, SIMAP provides an integrated clustering that is based on sequence homology as well as domain architectures. Clustering a large number of sequences by their pairwise sequence similarities is a non-trivial, computationally very expensive task. Among the many approaches that were successfully established, see e.g. (15), (16) or (17), the Tribe-MCL pipeline (18) provides the implementation of an efficient algorithm for large-scale detection of protein families based on the Markov cluster algorithm. Due to the huge number of pairwise similarities in SIMAP, even the application of the very fast Tribe-MCL pipeline requires preprocessing steps as described below. To avoid contamination of clusters by promiscuous domains as discussed in Ref. (17), we implemented a subclustering method that splits MCL clusters based on the domain architecture of the cluster members. Clusters are calculated using a hierarchical algorithm consisting of five main steps:
separation of sequences into the major taxonomic divisions—bacteria, archaea, eukaryota and viruses,
generation of non-redundant sets of sequences by pre-clustering of very similar sequences (ratios of alignment score between two sequences/maximal alignment score of the two sequences compared with itself and alignment/length must be both ≥90%),
Markov chain linkage clustering (18) of the similarity networks of non-redundant sequence sets into main clusters,
subclustering of the main clusters from Step 3 based on different domain architectures (more details on this method are given below) and
comparison of all member proteins of the main clusters from Step 3 between the taxonomic divisions to form metaclusters connecting related protein families from bacteria, archaea, eukaryota and viruses.
The cluster-centric view, which is available for all sets of protein shown in the SIMAP portal, allows exploring the similarity relations of the query protein and its homologs in an easy and convenient manner.
Search by similarity of domain architectures
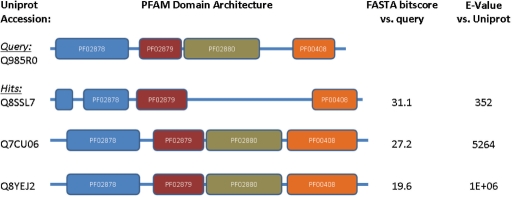
A novel search method in SIMAP addresses the task of finding homologs of multidomain proteins, especially in case of domain duplications or domain shuffling. The new ‘Domain similarity’ tool takes advantage of the consistent annotation of all sequences in SIMAP with their InterPro domains. Given a certain query protein, it allows to search for sequences of similar domain architecture. To quantitatively describe the evolutionary distance of two domain architectures—which is not trivial due to the specific evolution of multidomain proteins—we adapted a method proposed by Lin et al. (19). ‘Domain similarity’ searches are capable not only of refining the sort order of homologs, but also of finding remote homologs that lack sufficient sequence similarity for significant hits by FASTA and Smith–Waterman; however, their conservation is still detectable using position-specific scoring models (Figure 3).
Figure 3.
Example of remote homologs retrieved by the ‘Domain similarity’ tool of SIMAP. When searching the query sequence in the Uniprot database, high E-values result from the low bitscores. Thus, these proteins show insufficient pairwise sequence homology to the query and would not be found by database searches that are typically restricted to a maximal E-value of 10. However, the similar domain architectures suggest a common ancestry of these proteins.
Mapping of individual proteins into the public protein space
Due to the use of multiple identifiers for the same protein in different databases, an important but time-consuming task in bioinformatics is the transformation of a set of proteins into another domain of identifiers. This task is necessary also for proprietary databases that use special identifiers and should be mapped to recent public databases. A similar situation occurs for data from proteomics experiments.
SIMAP provides a very fast mapping between protein sets, based on the identity of protein sequences by comparison of their MD5 hashes.
In cases that do not allow for mapping by sequence identity, e.g. if sequences are fragmented or altered by unidentified residues, a more time-consuming mapping can be performed using PROMPT (20) that makes use of the SIMAP ‘SeqFinder’ function and provides the mapping by individual similarity searches.
FUTURE DIRECTIONS
In the future, the contents of the SIMAP database will be continuously updated every month to stay abreast of all published protein sequences. The recent statistics and information about contained databases can be found from the SIMAP web portal. Recently, the natural diversity of life and its underlying genetic information has been investigated by metagenomic projects. Sequences from environmental sequencing projects (‘metagenomes’) will be integrated into SIMAP soon. Together with future plans for the enhanced integration of functional annotations of proteins and the improvement of the clustering procedures, SIMAP will continue to facilitate individual discoveries as well as systematic downstream projects by providing a structured database of the pre-calculated sequence similarity and feature spaces.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the BOINCSIMAP community for donating their CPU power for the calculation of protein similarities and features and especially Jonathan Hoser for his continuous help in maintaining the BOINCSIMAP platform. The authors wish to thank SUN Microsystems Inc. for funding a fully equipped X4500 data center server, which is now hosting the SIMAP database, through a SUN Academic Excellence Grant. Funding to pay the Open Access publication charges for this article was provided by the GSF - Research Center for Enviromnent and Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold R, Rattei T, Tischler P, Truong MD, Stumpflen V, Mewes W. SIMAP—the similarity matrix of proteins. Bioinformatics. 2005;21:42–46. doi: 10.1093/bioinformatics/bti1107. [DOI] [PubMed] [Google Scholar]
- 6.Riley ML, Schmidt T, Artamonova II, Wagner C, Volz A, Heumann K, Mewes HW, Frishman D. PEDANT genome database: 10 years online. Nucleic Acids Res. 2007;35:D354. doi: 10.1093/nar/gkl1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 9.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Emanuelsson O, Nielsen H, Brunak S, Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 11.Rattei T, Walter M, Arnold R, Anderson DP, Mewes W. Using public resource computing and systematic precalculation for large scale sequence analysis. Lecture Notes Bioinformatics. 2007;4360:11–18. [Google Scholar]
- 12.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtz S. 2003. The Vmatch large scale sequence analysis software. Ref Type: Computer Program. 4-12-2003. [Google Scholar]
- 14.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kriventseva EV, Servant F, Apweiler R. Improvements to CluSTr: the database of SWISS-PROT+ TrEMBL protein clusters. Nucleic Acids Res. 2003;31:388–389. doi: 10.1093/nar/gkg035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan N, Sasson O, Inbar U, Friedlich M, Fromer M, Fleischer H, Portugaly E, Linial N, Linial M. ProtoNet 4.0: a hierarchical classification of one million protein sequences. Nucleic Acids Res. 2005;33:D216–D218. doi: 10.1093/nar/gki007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tetko IV, Facius A, Ruepp A, Mewes HW. feedback. 2005. Super paramagnetic clustering of protein sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin K, Zhu L, Zhang DY. An initial strategy for comparing proteins at the domain architecture level. Bioinformatics. 2006;22:2081. doi: 10.1093/bioinformatics/btl366. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt T, Frishman D. PROMPT: a protein mapping and comparison tool. BMC Bioinformatics. 2006;7:331. doi: 10.1186/1471-2105-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]