Abstract
The complete collection of evolutionary histories of all genes in a genome, also known as phylome, constitutes a valuable source of information. The reconstruction of phylomes has been previously prevented by large demands of time and computer power, but is now feasible thanks to recent developments in computers and algorithms. To provide a publicly available repository of complete phylomes that allows researchers to access and store large-scale phylogenomic analyses, we have developed PhylomeDB. PhylomeDB is a database of complete phylomes derived for different genomes within a specific taxonomic range. All phylomes in the database are built using a high-quality phylogenetic pipeline that includes evolutionary model testing and alignment trimming phases. For each genome, PhylomeDB provides the alignments, phylogentic trees and tree-based orthology predictions for every single encoded protein. The current version of PhylomeDB includes the phylomes of Human, the yeast Saccharomyces cerevisiae and the bacterium Escherichia coli, comprising a total of 32 289 seed sequences with their corresponding alignments and 172 324 phylogenetic trees. PhylomeDB can be publicly accessed at http://phylomedb.bioinfo.cipf.es
INTRODUCTION
A phylome is defined as the complete collection of phylogenies reconstructed for every single gene encoded in a genome (1). Although the term was coined several years ago, the development of high-quality, genome-wide collections of phylogenetic trees has been previously prevented due to large demands of time and computer power. Only recently, and thanks to new and faster algorithms and computers, the application of phylogenetics to whole genomes has become feasible. Large-scale phylogenetic studies provide very valuable information on the evolutionary relationships between genes of different species (2). Among other applications, the availability of complete phylomes can be exploited to map duplication and speciation events and thus infer orthology relationships (3), to determine the evolutionary relationships among taxa (4) and even to reconstruct ancestral metabolisms (5). Although some databases provide automatically derived and curated phylogenies (6–9), these follow a family-based approach, since they first group the genes into families and subsequently build a single phylogeny for each family. Moreover, the selection of species included is determined by the specific scopes of these databases. PhylomeDB provides phylomes reconstructed following a gene-based approach (3), in which the same high-quality phylogenetic pipeline is applied to each single gene encoded in a given genome. The resulting trees, alignments and tree-based orthology predictions can be easily accessed, queried and downloaded through a user-friendly web interface. In this article, the data content and web features of the first release of PhylomeDB are described.
DATABASE STRUCTURE AND CONTENT
General features
The current version of PhylomeDB contains the phylomes of three relevant organisms, including human and the two model species Saccharomyces cerevisiae and Escherichia coli. Future releases of PhylomeDB will incorporate phylomes for new species as well as novel versions of existing phylomes that may include different phylogenetic ranges or updated releases of their respective proteomes. To store all data associated with the phylomes, PhylomeDB uses a relational database. For each phylome, PhylomeDB provides: (i) a feature page, which contains general information on the proteomes included in the specific phylome as well as all the details of the phylogenetic pipeline used (e.g. http://phylomedb.bioinfo.cipf.es/index.html?Hsapiens001 for Hsapiens001 phylome); and (ii) an individual entry (Figure 1) for each protein encoded in the seed genome that provides the sequences, multiple sequence alignments and phylogenetic trees rendered by the phylogenetic pipeline. The database has been implemented in MySQL (http://www.mysql.com) but users can access all data via a user-friendly web interface that provides several query options (see below). To facilitate the interactivity, speed and functionality of the website, the web interface has been developed in Asynchronous JavaScript and XML (AJAX) technology. Although the use of the web interface of PhylomeDB is quite intuitive, a user's manual is provided in the form of a wiki page (http://www.mediawiki.org). This provides an easy mechanism by which registered users can help PhylomeDB developers correcting and expanding the documentation.
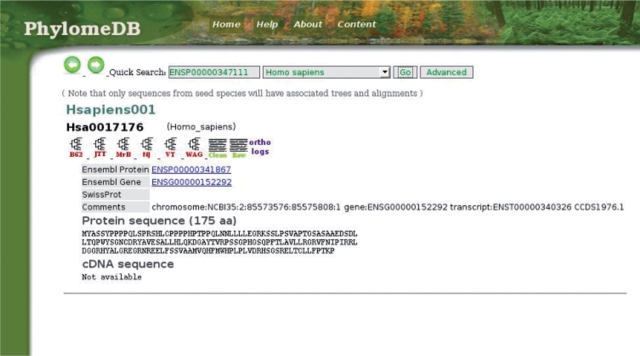
Figure 1.
Screenshot of a PhylomeDB entry page. The entry page of an individual seed protein (Hsa0017176) of the human phylome (Hsapiens001), includes information on the sequence and links to (i) the feature page containing all necessary information of the phylogenetic pipeline used; (ii) all the phylogenetic trees derived by the different programs and models used in the pipeline, in this case a NJ tree, ML trees derived from JTT, WAG, RtREV and BLOSUM62 models, and the consensus tree derived from the Bayesian analyses; (iii) the raw and trimmed (clean) alignments and (iv) the list of tree-based orthology predictions for the species included in the tree.
Phylome reconstruction pipeline
All phylomes included in PhylomeDB have been generated following a similar high-quality phylogenetic pipeline that is described more extensively elsewhere (3). The particularities of each phylome as well as the list of species included are comprehensively described in the corresponding feature pages that can be accessed by clicking on the phylome code (e.g. Hsapiens001). Each phylome is defined by the proteome of the seed species and a specific dataset of proteomes, including several other species. The proteomes are downloaded from sequence databases such as Ensembl, EBI and those from specific genome sequencing projects, details of the source of the sequences are provided in the feature pages of the phylomes. In summary, the pipeline proceeds as follows: for each protein of the seed proteome, a Smith–Waterman (10) search is performed against the corresponding proteome dataset to retrieve a set of proteins with a significant similarity (e-value <10−3). Only sequences that align with a continuous region longer than 50% of the query sequence are selected as homologs. These sets of homologous sequences are subsequently aligned using MUSCLE 3.6 (11) with default parameters. Positions in the alignment with gaps in more than 10% of the sequences are removed, unless such procedure removes more than one-third of the positions in the alignment. In such cases, the percentage of sequences with gaps allowed is automatically increased until at least two-thirds of the initial positions are conserved. Phylogenetic trees are derived from the resulting alignments by using several methods, which may include: (i) Neighbor Joining (NJ) trees using scoredist distances as implemented in BioNJ (12); (ii) Maximum Likelihood (ML) as implemented in PhyML v2.4.4 (13) assuming a discrete gamma-distribution model with four rate categories and invariant sites, where the gamma shape parameter and the fraction of invariant sites are estimated from the data only in the case of the human phylome and (iii) Bayesian phylogenetic reconstruction using Mr Bayes for 100 000 generations in two rounds of two chains each (14). Since, both ML and Bayesian analyses are model-based approaches that can provide divergent results when different evolutionary models are assumed. In such cases, all phylogenetic trees derived from the different models are provided and the model best fitting the data, as judged by the AIC criterion (15), is indicated. The models used in the different phylomes are listed in Table 1.
Table 1.
Current content in phylomeDB
| Phylome code | Seed species | Seed proteins | Species content | Total trees | Phylogenetic methods | Brief description |
|---|---|---|---|---|---|---|
| Hsapiens001 | Homo sapiens | 21 588 | 38 | 157 233 | NJ, Bayesian ML(JTT,WAG,B62, RtREV, MtREv) | 38 eukaryotic species from Ensembl, Integr8 and 3 other sources. |
| Ecoli001 | E. coli | 4604 | 421 | 9280 | NJ,ML(JTT,WAG) | 421 eukaryotic, archaeal and bacterial species from Integr8. |
| Scerevisiae001 | S. cerevisiae | 5811 | 421 | 5811 | NJ,ML(JTT) | The same species set as Ecoli001 |
| Total | 32 003 | 443 | 172 324 |
For each phylome included in the current release of PhylomeDB, the PhylomeDB internal code, the number of seed proteins, the number of species included, the total number of phylogenetic trees, the phylogenetic reconstruction methods and a brief description is provided. Phylogenetic methods are indicated as follows: Neighbor Joining (NJ), Bayesian analysis (Bayesian) and Maximum Likelihood (ML), which can be performed using JTT, WAG, Blosum62 (B62), RtREV and MtREV evolutionary models. Bayesian analysis was always performed using the evolutionary model that rendered the best likelihood in the ML analysis.
Data formats
Inspired by the success of the information standard for microarray analyses (MIAMA), the need for a similar minimum information about a phylogenetic analysis standard (MIAPA) has been suggested (16). Although the developing of MIAPA standards is still on progress, several general guidelines have been proposed (16). In accordance to these guidelines, PhylomeDB provides comprehensive information on the programs and parameters used for each step of the pipeline so that the phylogenetic reconstruction can be reproduced. Since the same phylogenetic pipeline is applied to all seed proteins in a phylome, such details are provided in the features page of the corresponding phylome. Moreover, all alignments and trees rendered by the phylogenetic pipeline are provided in standard newick and phylip formats, respectively. As soon as new guidelines and MIAPA standards are developed, these will be implemented in PhylomeDB. Besides phylogenetic trees and alignments, PhylomeDB provides tree-based orthology and paralogy predictions. These predictions are generated from the seed sequence by mapping duplication and speciation events on the tree as determined by a species-overlap algorithm (3). In contrast to alternative, phylogeny-based methods that use reconciliation of the gene tree with the species tree, the PhylomeDB algorithm uses the level of overlap in the species connected to two related nodes to decide whether their parental node represents a duplication or speciation event. Briefly, the algorithm visits all nodes that connect the seed protein to the root of the tree and marks it as a duplication event if one or more species are shared by its two children nodes.
When the orthology prediction algorithm is launched, phylomeDB displays the list of predicted orthologs and paralogs for the seed sequence, as well as a tree with the corresponding speciation and duplication edges indicated in red and blue (Figure 3A), respectively, are provided. The full set of predicted orthology and paralogy relationships for each phylome can be downloaded from the download section of the database.
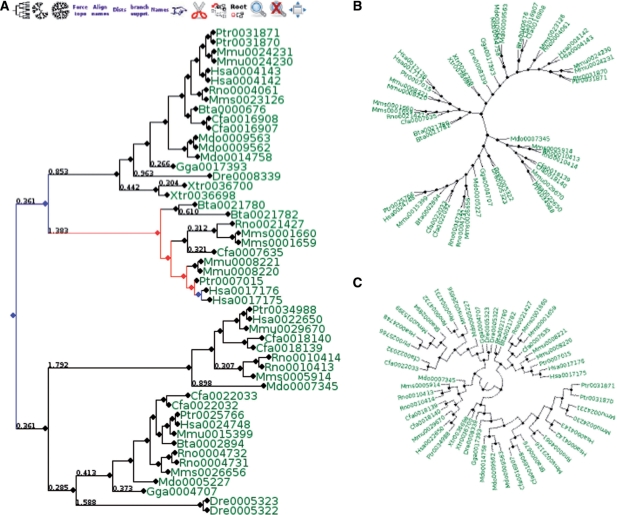
Figure 3.
Visualization of phylogenetic trees: different views of the ML phylogenetic tree, using JTT evolutionary model, for the seed protein (Hsa0017176), as displayed by the environment for tree exploration tool (ETE). Trees can be represented in rectangular (A), circular (B) or radial (C) modes. Several information labels such as support values or branch distances (A) can be displayed on nodes and edges, which can also be colored to indicate different evolutionary event. In (A) an example is shown where the lineages leading to the seed protein (Hsa0017176) are colored in blue or red to mark duplications and speciation events, respectively. This feature is shown after running the orthology prediction algorithm by clicking on the ‘orthologs’ icon. Using the ETE toolbar (icons on the top of the section A), the user can navigate within the tree and browse it using different visualization options.
All protein sequences included in PhylomeDB are given a unique alpha-numeric ID (PhylomeID), which includes a three-letters code designating the species, followed by a sequential number. Codes for the 443 organisms included in PhylomeDB are listed in the features page of the corresponding phylomes. Correspondence between PhylomeIDs and IDs from external databases such as Swissprot and Ensembl are given in the corresponding sequence entries. Moreover, an ID converter tool provides the equivalences between Phylome ID and other selected IDs.
DATABASE ACCESS AND WEB FEATURES
Browsing and querying
PhylomeDB is publicly accessible at http://phylomedb.bioinfo.cipf.es. There are various ways in which users can access data stored in PhylomeDB. The database can be browsed by selecting a specific phylome from the home or content pages. In this case, a table appears that lists the entries to all the seed proteins included in the phylome. By clicking in a specific PhylomeID, the corresponding entry is shown. Each entry contains information on the seed protein and the homologous proteins included in the phylogenetic analysis together with the corresponding alignments and phylogenetic trees.
Moreover, an ID text-based search is available for users to search for specific proteins by their PhylomeID, Swissprot or Ensembl IDs, among others, and, finally, the user can search using a specific protein sequence. In such case, a Smith–Waterman search is performed against all seed proteins included in the database and the significant hits (e-value = 10−3), with links to their entry pages, are displayed.
Visualization
PhylomeDB provides all alignments, sequences and trees in separate files in a plain text format, so that the files can be downloaded and visualized with the user's favorite tool. Moreover, PhylomeDB incorporates multiple sequence alignment and tree visualization tools to facilitate the online visualization of the data. Alignments can be visualized with Jalview (Figure 2), which is a java application enabling fast viewing of large multiple sequence alignments (17). PhylomeDB implements a web plugin of the program ETE (Environment for Tree Exploration) developed by J. Huerta-Cepas. This program is an interactive tree viewer that implements different visualization, search and browse modes (Figure 3). Some of its most interesting features include the visualization of trees using rectangular, circular or radial representation modes, rooting options and functions to collapse and/or extract sub-trees. Selected (sub) tree visualization can be downloaded as standard PNG images. A complete description about the ETE interface can be accessed through the PhylomeDB user's manual page.
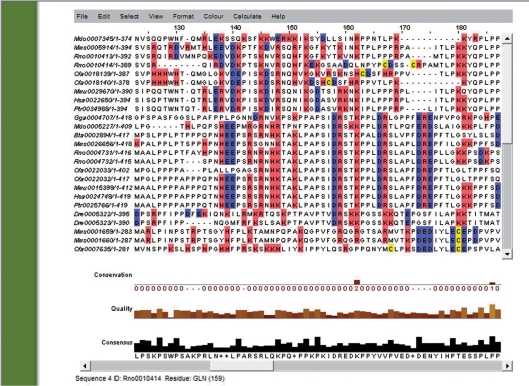
Figure 2.
Visualization of multiple sequence alignments: Example of the trimmed (clean) multiple sequence alignment of the example entry (Hsa0017176) as displayed by JalView (17). Sequence names are indicated by PhylomeID names, which include the three-letters species code followed by a number. The levels of sequence conservation and quality of each column, as well as a consensus sequence are indicated bellow. Colors, sequence format and visualization options can be changed using JalView interface.
PhylomeDB entries can be easily linked to from external pages using the following URL scheme: http://phylomedb.bioinfo.cipf.es/index.html?Hsapiens001&Hsa0000001 to link to the entry Hsa0000001 of Hsapiens001 phylome. In this manner, protein databases not specifically focused in phylogenetic information can provide links to the phylogenies of their proteins.
FUTURE PERSPECTIVES
In summary, PhylomeDB has been developed as a database for storing and querying complete collections of gene phylogenies for complete genomes. We are continuing to expand the PhylomeDB database to incorporate data from other model species using the pipeline described above. We encourage other groups to contact us in order to suggest new phylomes to be generated or to submit phylomes generated with similar procedures. In any case, only phylomes that have been reconstructed following similar high-quality procedures will be stored in PhylomeDB. New enhancements that we are focusing on for the short- to medium-term include the following: (i) increase links to other external databases providing additional information such as functional annotation; (ii) provide enriched newick format with information on the nodes such as labels for speciation and duplication events and (iii) implement a topology search tool so that trees can be searched for the presence of a given topology. A new release of PhylomeDB is expected on a yearly basis.
ACKNOWLEDGEMENTS
J.H.-C. is supported by a grant from the Fundación Genoma España and the Instituto Nacional de Bioinformática. T.G. is recipient of a post-doctoral fellowship from the European Molecular Biology Organization (EMBO LTF 402-2005) and of an ISCIII Grant from the Spanish Ministry of Health (06/00213). A fraction of the phylogenetic trees stored in PhylomeDB was reconstructed using the super-computer Mare Nostrum, from the Barcelona Supercomputing Centre. We are grateful to J. Burguet for providing technical support and to L. Arbiza for critically reading the manuscript. Funding to pay the Open Access publication charges for this article was provided by ISCIII Grant from the Spanish Ministry of Health (06/00213).
Conflict of interest statement. None declared.
REFERENCES
- 1.Sicheritz-Ponten T, Andersson SG. A phylogenomic approach to microbial evolution. Nucleic Acids Res. 2001;29:545–552. doi: 10.1093/nar/29.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabaldón T. Evolution of proteins and proteomes, a phylogenetics approach. Evol. Bioinform. Online. 2005;1:51–56. [PMC free article] [PubMed] [Google Scholar]
- 3.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldon T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comas I, Moya A, Gonzalez-Candelas F. From phylogenetics to phylogenomics: the evolutionary relationships of insect endosymbiotic gamma-Proteobacteria as a test case. Syst. Biol. 2007;56:1–16. doi: 10.1080/10635150601109759. [DOI] [PubMed] [Google Scholar]
- 5.Gabaldón T, Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301:609. doi: 10.1126/science.1085463. [DOI] [PubMed] [Google Scholar]
- 6.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Dickerman AW. GeneTrees: a phylogenomics resource for prokaryotes. Nucleic Acids Res. 2007;35:D328–D331. doi: 10.1093/nar/gkl905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duret L, Mouchiroud D, Gouy M. HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 13.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 14.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 15.Akaike H. Proceedings of the 2nd International Symposium on Information Theory; Budapest, Hungary. 1973. pp. 267–281. [Google Scholar]
- 16.Leebens-Mack J, Vision T, Brenner E, Bowers JE, Cannon S, Clement MJ, Cunningham CW, dePamphilis C, deSalle R, et al. Taking the first steps towards a standard for reporting on phylogenies: minimum information about a phylogenetic analysis (MIAPA) Omics. 2006;10:231–237. doi: 10.1089/omi.2006.10.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]