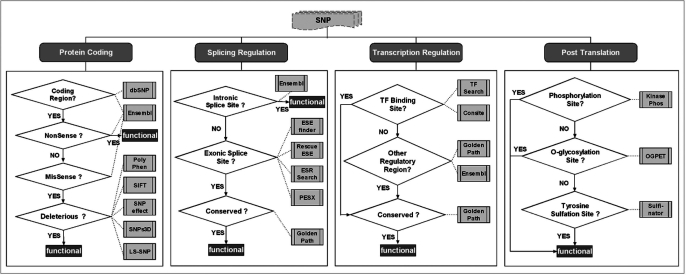
Figure 1.
Decision procedure for functional SNP assessment. Each SNP is examined for deleterious effects with respect to each functional category (i.e. protein coding, splicing regulation, transcriptional regulation and post-translation—as shown in the top part of the figure). For each category, a series of tests is executed to determine whether the SNP has a functional impact. First the type (coding, intronic, etc.) of the genomic region is identified, using data from dbSNP (3) and Ensembl (4). Once this is determined, other tests are performed. For example, to assess if a SNP has a deleterious effect on protein coding, it first must be located on a coding region. Ensembl (4) is used to examine if this is a nonsense mutation, in which case the SNP is considered to be deleterious. Otherwise—if the SNP is a missense mutation, it is further tested by five different tools [PolyPhen (6), SIFT (7), SNPeffect (8), SNPs3D (9) and LS-SNP (10)] to check if the non-synonymous substitution is deleterious. A majority vote among these tools concludes the process, and identifies the SNP as either having a potentially deleterious functional impact (denoted ‘functional’ in the figure) or not.