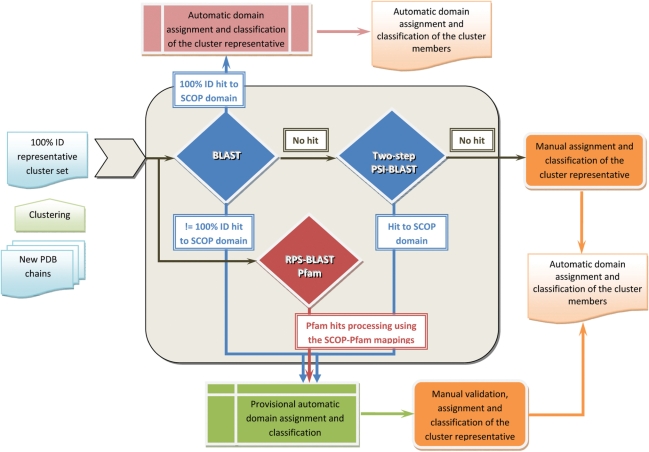
Figure 1.
Workflow of the SCOP update protocol. The update sequence set of new unclassified structures is derived from the PDB SEQRES record. Disordered regions at the termini are masked. The update sequences are clustered using a threshold of 100% identity and 95% coverage for the inclusion of protein sequence into the cluster set. The resulting clusters are used to select a representative sequence set. This dataset is used as a primary input to the pre-classification pipeline. The representative cluster set is first compared using BLAST against itself and a database of non-redundant representative ASTRAL sequences for SCOP domains. This step allows detection of close homologs, usually members of the same SCOP family. Representative sequences without significant sequence match (E-value = 0.001) are further used for two-step PSI-BLAST searches. In the first step, a position-specific scoring matrix (PSSM) is generated by searching the NCBI non-redundant protein database. The resulting PSSM is saved after ten PSI-BLAST iterations or less if the program converges. In the second step, each saved PSSM is used to scan databases of representative ASTRAL and update sequences. In addition, the representative cluster set of unclassified proteins is submitted for RPS-BLAST search against a database of Pfam profiles. The resulting matches are then compared with the matches of pre-computed large-scale comparisons of SCOP domains and Pfam families. A provisional SCOP classification assignment is made for those proteins with a matching region in Pfam that has given a hit to SCOP domain. The results of both RPS-BLAST and PSI-BLAST are used to identify relationships between more distant homologs that are likely to be members of the same SCOP superfamily. Update proteins that are identical or nearly identical to domains classified in the current SCOP release or in the SCOP developmental version are classified automatically. The remaining proteins with and without provisional classification are curated manually.