Figure 1. Use of HMMs to identify DHCs encoded in the genomes of 24 diverse eukaryotes.
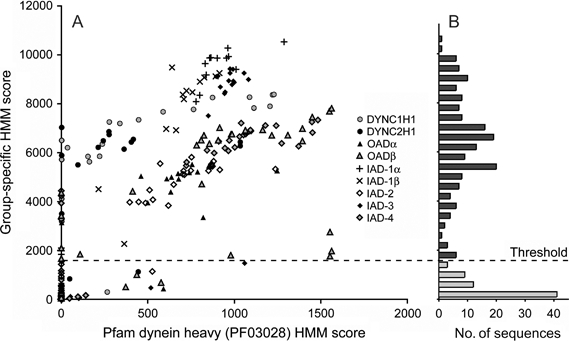
A) Performance of the Pfam dynein heavy HMM, PF03028.5, against group-specific HMMs. The y-axis shows score for group-specific HMM giving highest-scoring match. All predicted polypeptides in the 24 genomes with a score >0 on either axis are shown. B) Histogram of the distribution of matches to group-specific HMMs used to define the dynein heavy dataset (score > threshold). On the basis of the distribution, a liberal threshold was chosen to encompass the vast majority of dynein-like sequences without the inclusion of excessive numbers of false positives and extremely divergent sequences that accumulate in the low-score tail.
