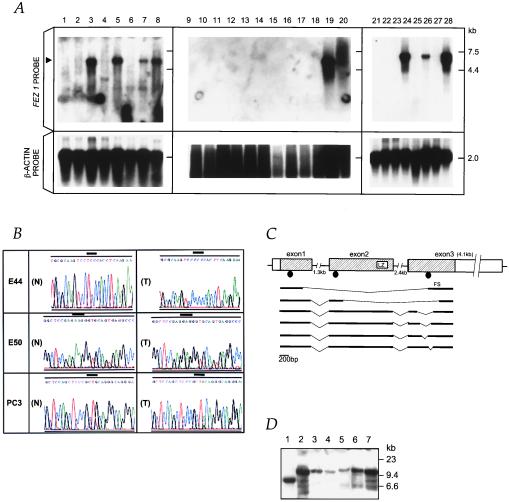
Figure 3.
Alterations of FEZ1 in tumors. (A) FEZ1 gene expression in cancers. Probes used were a FEZ1 cDNA probe (Upper) and a control β-actin probe (Lower). Poly(A)+ RNA (5 μg) from cancer cell lines was loaded. The arrowhead on the left indicates the 6.8-kb transcript of FEZ1. RNAs were from: esophageal cancer cell lines KYSE170 (lane 1), TE12 (lane 2), TE8 (lane 3), and TE3 (lane 4); prostate cancer cell lines DU145 (lane 5), LNCaP (lane 6), PC3 (lane 7), and normal prostate (lane 8); breast cancer cell lines MB231 (lane 9), SKBr3 (lane 10), BT549 (lane 11), HBL100 (lane 12), MB436 (lane 13), BT20 (lane 14), MB543 (lane 15), MB175 (lane 16), MCF7 (lane 17), and T47B (lane 18); normal breast (lane 19); and total RNA of normal breast (lane 20); promyelocytic leukemia cell line HL60 (lane 21); cervical cancer cell line HeLa S3 (lane 22); chronic myelogenous leukemia cell line K562 (lane 23); lymphoblastic leukemia cell line MOLT4 (lane 24); Burkitt’s lymphoma cell line Raji (lane 25); colorectal adenocarcinoma cell line SW480 (lane 26); lung cancer cell line A549 (lane 27); and melanoma cell line G361 (lane 28). (B) Sequence chromatograms of three mutations. A point mutation (TCC/Ser → CCC/Pro) at codon 29 was found (T) in an primary esophageal cancer, E44. As controls, normal sequences from normal DNA from patient E44 and BAC sequence (N) were analyzed, both producing the same result. A bold line indicates the altered codon. In primary esophageal cancer, E50, a point mutation (AAG/Lys → GAG/Glu) at codon 119 was found (T). The normal BAC sequence is shown in (N). The point mutation (CAG/Gln → TAG/STOP) at codon 501 was observed in prostate cancer PC3 cells (T). Note that the sequence chromatogram from this cell line is shown in the 3′ to 5′ direction. Repeated sequencing showed a weak signal corresponding to guanine (G) within a large adenine (A) signal in the first nucleotide at codon 501, suggesting the retention of the normal allele in a fraction of the cancer cells. Expression analysis with RT-PCR detected only the mutated transcript in this cell line. The normal BAC sequence is shown in (N). (C) Truncated FEZ1 transcripts observed in cancers. The normal exon/intron structure (top drawing) was determined by sequencing of brain, prostate, and esophagus cDNAs and by sequencing FEZ1 gene in BAC. The boxes indicate exons, and the shaded areas show the ORF of 1,788 bp. The introns are depicted by horizontal lines. Aberrant transcripts observed in tumors are depicted in bold lines. The location of mutations are shown as closed circles. LZ, leucine-zipper motif; FS, frameshift. (D) Southern blot analysis of the FEZ1 gene locus. High-Mr DNAs from cancer cells were cleaved with EcoRI, separated electrophoretically, transferred to a nylon membrane, and probed with the 1.7-kb FEZ1 ORF probe. DNAs (10 μg/lane) were from lanes: 1, MB436S; 2, normal; 3, MB231; 4, MB361; 5, TE8; 6, TE3; 7, normal (different origin from normal DNA in lane 2).