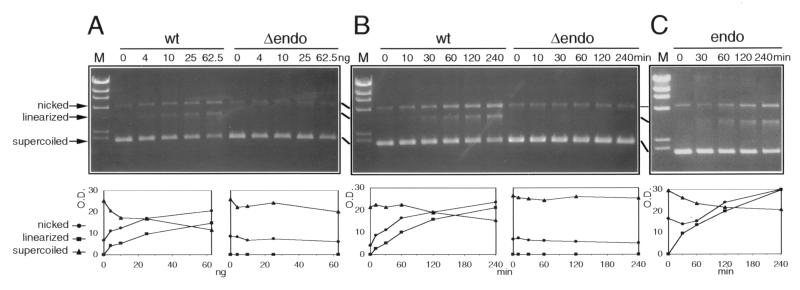
Figure 4.
Endonuclease activity of MED1. (A) The indicated increasing amounts of wild-type MED1 (wt) and a deletion mutant lacking the endonuclease domain (Δendo) were incubated with 0.25 μg of supercoiled pCR2 plasmid at 37°C for 30 min. Reaction products were analyzed by agarose gel electrophoresis (Upper). The migration of nicked, linearized, and supercoiled DNA is indicated. A small amount of nicked molecules is present in the plasmid DNA substrate at time zero. M, λ/HindIII digest size standards. (Lower) Densitometric analysis of the nicked (circles), linearized (squares), and supercoiled (triangles) bands. OD, in thousands of units. (B) Time course of MED1 endonuclease activity. MED1 wt and Δendo (12.5 ng, approximately 170 fmol) were incubated with 1.5 μg (approximately 580 fmol) of pCR2 plasmid at 37°C; at the indicated time points, an aliquot of the reaction was removed for electrophoretic analysis. (C) Endonuclease activity of the recombinant MED1 endonuclease domain (codons 455–580, endo). Endo (3.6 ng, approximately 170 fmol) was incubated with 1.5 μg (approximately 580 fmol) of pCR2 plasmid at 37°C and processed as in B.