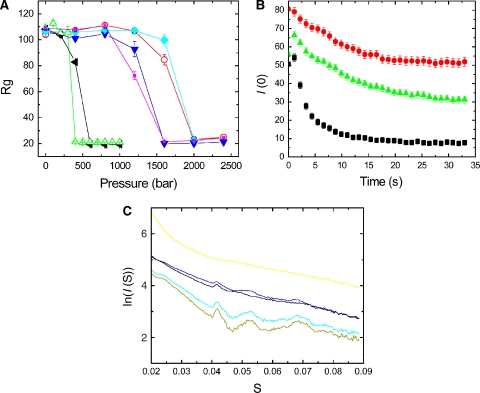
Figure 3.
(A) The radii of gyration of the complexes obtained from SAX patterns are plotted as a function of static pressure. ParM-GTP (green) and ParM-ATP (black) depolymerize at pressures of 400 and 500 bar, respectively, whereas ParM-GTPγS (pink), ParM-GMPPNP (blue), ParM-ATPγS (cyan), and ParM-AMPPNP (orange) disintegrate at much higher pressures. The protein concentration was 10 mg/ml (B) Following a rapid pressure jump (1 → 400 bar), the forward scattering intensity was monitored over time. ParM-GTP filaments (black) depolymerize much sooner than ParM-Mix (green) and ParM-ATP filaments (red). (C) SAX patterns from ParM in the presence of various nucleotides. ParM-GMPPNP (light green) and ParM-AMPPNP filaments (cyan) have the highest polymer content, with predominant peaks noticeable. The sharp large first peak arises from the 24 Å subunit repeat. ParM-ATP (brown) and ParM-GTP filaments (blue) have less helical content and more monomers present, indicative by the weakening of the 24 Å and higher-order peaks and an increase in diffusive background. ParM even at high concentrations of 10 mg/ml did not polymerize in the presence of GDP, giving a SAX pattern typical for a monomer (yellow).