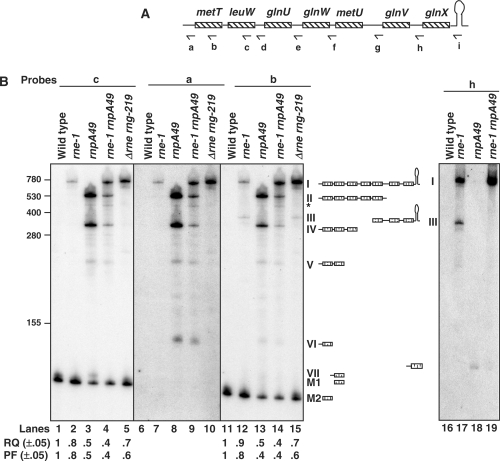
Figure 6.
Analysis of processing of the metT polycistronic transcript. (A) Schematic presentation of metT operon (not drawn to scale). Relative positions of the oligonucleotide probes (a: METT-UP, b: METT-265, c: LEUW-338, d: GLNU-UP, e: GLNW-UP, f: METU-UP, g: GLNV-UP, h: GLNX-UP and i: GLNX-TER) used in the northern analysis are shown below the cartoon. (B) Northern analysis of metT leuW glnU glnW metU glnV glnX transcripts. Total RNA (12 μg/lane) was separated on 6% PAGE, transferred to a nylon membrane and probed multiple times with probes a–i (A) as described in Materials and Methods section. The genotypes of the strains used are noted above each lane. The deduced structures and the names for the processing intermediates are shown. The weak band marked with a asterisk (*) is due to either a non-specific or weak cleavage by an unidentified ribonuclease in the middle of the metU-coding sequence, since it hybridized to the probes a–f (A and B, lanes 1–15, data not shown) but not to the probes g–i (B, lanes 16–19, data not shown). RQ and PF of the mature tRNAMet (M2) and tRNALeu2 (M1) were calculated as described in Figure 1. The RNA size standards (nucleotide) (Invitrogen) are shown to the left. Mature tRNALeu3 (M1) is 85 nt in length while the mature tRNAMet (M2) is 79 nt.