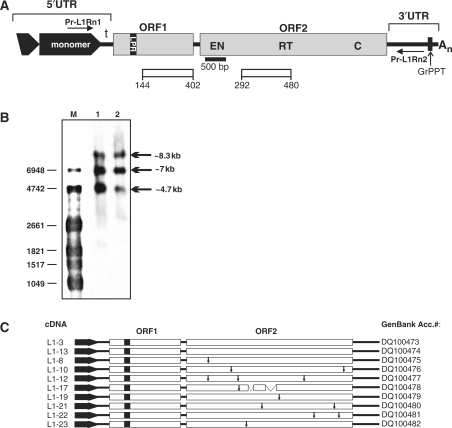
Figure 1.
Full-length L1Rn elements are transcribed in RCL cells. (A) Structure of L1Rn. A functional full-length L1Rn element is characterized by two ORFs flanked by 5′ and 3′ UTRs. The bipartite 5′ UTR consists of a monomer, which can be tandemly repeated, and a non-repeated tether (t). The most 5′ monomer is only partially duplicated (black truncated box with arrowhead) in all genomic rat elements identified so far. Horizontal arrows indicate the binding sites of the oligonucleotides Pr-L1Rn1 and Pr-L1Rn2 used to amplify cDNAs generated from full-length transcripts. The binding site of the 500-bp digoxigenin-labeled probe used to detect L1-specific transcripts is localized at the 5′ end of ORF2 (black bar). Open bars represent ORF1- and ORF2-encoded polypeptides against which monoclonal antibodies were raised. The polypeptides are covering amino acid positions 144–402 and 292–480 of ORF1p and ORF2p, respectively (accession no. DQ100480). LPR, length polymorphism region; GrPPT, G-rich polypurine tract; An, A-rich 3′ tract; EN, endonuclease; RT, reverse transcriptase; C, cysteine-rich motif. (B) L1Rn transcriptional products in RCL cells. PolyA+ RNA was isolated from RCL cells that had reached the maximum population density of ∼106 cells/ml (lane 2) and from cells that were UV-irradiated (lane 1). Two microgram of each RNA were separated by agarose gel electrophoresis and subjected to northern blot analysis using a 500-bp probe (Figure 1A). (C) Schematic structures of 10 L1Rn cDNAs synthesized from poly(A)+ RNA from UV-irradiated RCL cells. cDNAs are flanked by primer sequences Pr-L1Rn1 and Pr-L1Rn2. Names of the resulting cDNAs are listed on the left, while accession numbers are specified on the right. Termination codons within ORF2 sequences are indicated by vertical arrows. Two deletions in the ORF2-coding region of cDNA L1-17 covering 103 and 220 nts, respectively, are indicated by interrupted bars.